Precision Folds
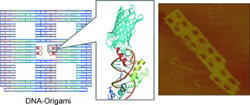
DNA is not merely a carrier of genetic information; DNA is a useful building material for nanoscale structures. In a way similar to origami, the Japanese art of paper folding, a long single strand of DNA can be folded into nearly any three-dimensional shape desired with the use of short DNA fragments.
The DNA nanostructure can also be equipped with specific docking sites for proteins. In the journal Angewandte Chemie, a team led by Takashi Morii at the University of Kyoto (Japan) has now introduced a new method for attaching the proteins by means of special “adapters” known as zinc-finger proteins.
Physiological processes and chemical reactions in cells are highly specific and take place in several reaction steps. Multiple enzymes must cooperate in order to catalyze the sequential steps of the required chemical transformations – and are much more efficient at it than synthetic systems. The natural systems can only be effectively imitated if the individual enzymes and factors have the correct relative orientations in space. DNA-origami structures can be used as “molecular switchboards” to arrange enzymes and other proteins with nanometer-scale precision.
Various methods for binding proteins to DNA-origami structures have previously been developed, but in most cases they require modification of the protein. “A method based only on proteins is desirable,” says Morii, “because it would simplify and accelerate the binding of proteins to the origami.”
Morii and his team settled on the use of zinc-finger proteins as “adapters”. A polypeptide chain of zinc-finger protein grabs a zinc ion to form a stable compact fold; this fold referred to as a “zinc finger” and can bind to specific DNA patterns. It is possible to make zinc fingers that recognize any DNA pattern desired.
The scientists produced rectangular origami structures with several defined cavities. At these locations, the origamis contain various DNA-recognition patterns for different zinc fingers. The researchers then made proteins that contain zinc-finger units at one end and a fluorescing protein or biotin molecule at the other end. Biotin binds specifically to the large protein streptavidin. Atomic force microscopic images show that the streptavidin molecules always bind specifically to the intended cavity in the origami rectangle.
“Our results demonstrate that zinc fingers are suitable site-selective adapters for targeting specific locations within DNA-origami structures,” says Morii. “Several different adapters carrying different proteins can independently bind at defined locations on this type of nanostructure.”
About the Author
Dr Takashi Morii is a Professor at Kyoto University with appointments at the Institute of Advanced Energy. His main specialty are bioorganic chemistry, chemical biology, and the development of technologies in constructing functional biomacromolecules.
Author: Takashi Morii, Kyoto University (Japan), http://akweb.iae.kyoto-u.ac.jp/material/en/index.html
Title: Zinc-Finger Proteins for Site-Specific Protein Positioning on DNA-Origami Structures
Angewandte Chemie International Edition, Permalink to the article: http://dx.doi.org/10.1002/anie.201108199
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…
Making diamonds at ambient pressure
Scientists develop novel liquid metal alloy system to synthesize diamond under moderate conditions. Did you know that 99% of synthetic diamonds are currently produced using high-pressure and high-temperature (HPHT) methods?[2]…

Eruption of mega-magnetic star lights up nearby galaxy
Thanks to ESA satellites, an international team including UNIGE researchers has detected a giant eruption coming from a magnetar, an extremely magnetic neutron star. While ESA’s satellite INTEGRAL was observing…





















