Nanotech instruments allow first direct observations of RNA ’proofreading’
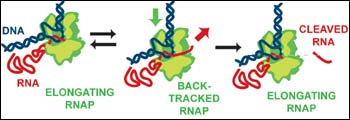
The RNA polymerase proofreading mechanism (Credit: E.. Abbondanzieri) <br>
When Ralph Waldo Emerson said that nature pardons no mistakes, he wasn’t thinking about RNA polymerase (RNAP) – the versatile enzyme that copies genes from DNA onto strands of RNA, which then serve as templates for all of the proteins that make life possible.
Emerson’s comment notwithstanding, RNAP makes plenty of mistakes but also proofreads and corrects them before they have a chance to create abnormal proteins. The error-prone nature of RNAP is not surprising given the size of its task. In human cells, for example, the RNAP enzyme has to make precise genetic copies from a DNA double helix that consists of billions of chemical bases known as A, T, G and C. It works like this: After latching onto the double helix, RNAP pulls it apart and starts building new RNA molecules by copying one DNA base at a time.
With thousands of A’s, T’s, G’s and C’s to transcribe, RNAP sometimes gets confused and copies the wrong base. Such errors occur roughly once every 1,000 bases, but RNAP’s remarkable self-correcting mechanism manages to catch most of them.
’’If the error is allowed to propagate, it could result in a bad protein or a wrong protein, but RNAP is an incredibly smart enzyme,’’ says Steven M. Block, a professor of biological sciences and of applied physics at Stanford University. ’’When RNAP adds the wrong base, it backs up on the DNA helix a little bit, cleaves off the piece of RNA that has the bad base in it and starts up again. That’s the hypothesis, at least.’’
New experiments
In a new study in the journal Nature, Block and his colleagues present strong evidence to support this proofreading hypothesis. Their results – based on actual observations of individual molecules of RNAP – are posted on Nature’s website: http://www.nature.com. In another set of experiments published in the Nov. 14 issue of Cell magazine, the researchers discovered that RNAP makes thousands of brief pauses as it pries open and copies the DNA double helix.
’’Together these two papers push the study of single proteins to new limits,’’ Block said. ’’We’ve been able to achieve a resolution of three angstroms – the width of three hydrogen atoms – in our measurements of the progress of this enzyme along DNA. In so doing, we’ve been able to visualize a backtracking motion of just five bases that accompanies RNAP error-correction or proofreading.’’
Both studies were conducted using two-dimensional optical force clamps – unique instruments designed and built by the Block lab. Located in soundproofed and temperature-controlled rooms in the basement of Stanford’s Herrin labs, these devices allow researchers to trap a single molecule of RNAP in a beam of infrared light, and then watch in real time as it moves along a single molecule of DNA.
’’We’ve been able to reduce drift and noise in our instruments to such an extent that we can see the tiniest motions of these molecules, through distances that are less than their own diameters,’’ Block explained. ’’Studying one macromolecule at a time, you learn so much more about its properties, but these kinds of experiments were just pipedreams 15 years ago.’’
Stops and starts
In their experiments, the Block team conducted more than 300 observations of single RNAP molecules extracted from E. coli bacteria. Although structurally somewhat different from human RNAP, the E. coli enzyme plays a very similar role in the complex transfer from gene to RNA to protein.
Using the optical clamp, researchers found that RNAP does not move at a steady pace along the DNA double helix but rather undergoes a fitful series of unexplained starts and stops. ’’This enzyme is either full on or full off, as far as we can tell,’’ Block said. ’’It moves at about ten to 15 bases a second and pauses on average about once every 100 bases. Pausing is ubiquitous. About 97 percent are short pauses that last between 1.5 to 4.5 seconds. The other three percent are long pauses – from 20 seconds to over 30 minutes.’’
The study published in Cell focused on the short pauses, he added: ’’What we learned is that short pauses do not involve backtracking. What are these pauses? The answer is we don’t know.’’
The Nature study looked at the long pauses, which the researchers discovered only occur during backtracking – the hypothetical proofreading event during which RNAP is believed to slide backwards and snip off defective RNA. To test the hypothesis, the scientists added two proteins called GreA and GreB that are known to speed up the RNA cleaving process in E. coli. It turned out that adding GreA and GreB significantly shortened the backtracking pauses, providing strong evidence that backtracking and proofreading go hand in hand.
Medical consequences
Finding the answer could have significant ramifications for biology and for human health, observed physics graduate student Joshua W. Shaevitz, co-lead author of the Nature study.
’’When it comes to transcribing genetic code from DNA to RNA, fidelity is important,’’ he said. ’’If the protein comes out wrong, it may be lethal to the cell or to the organism.’’
Certain antibiotics are known to increase the error rate during translation from RNA to protein, added applied physics graduate student Elio A. Abbondanzieri, co-lead author of the Nature paper and co-author of the Cell study.
’’In the future we hope to study RNAP as it backs up in other situations when there is no mistake,’’ he said.
’’This research allows us to see a process essential to life at a level of detail never before possible,’’ said Catherine Lewis, chief of the biophysics branch in the National Institute of General Medical Sciences (NIGMS), which funded both studies. ’’It’s analogous to measuring the speed and direction of a single car, while other studies saw only the rush of traffic. This basic research will advance our understanding of how errors in transcription underlie disease and will pave the way for better tools to address such problems.’’
Other co-authors of the Cell study are Keir C. Neuman, a postdoctoral fellow in Stanford’s Department of Biological Sciences; Robert Landick of the University of Wisconsin; and Jeff Gelles of Brandeis University. Landick also co-authored the Nature study.
COMMENT: Steven M. Block, Departments of Biological Sciences and Applied Physics: 650-724-4046, sblock@stanford.edu
EDITORS: The study, ’’Backtracking by single RNA polymerase molecules observed at near-base-pair resolution,’’ is available on Nature magazine’s Website,http://www.nature.com. ’’Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking’’ is published in the Nov. 14 edition of the journal Cell. Additional information, videos and other images are available online at http://www.stanford.edu/group/blocklab/NatureBacktracking/.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















