Scientists Reveal a New Way Viruses Cause Cells to Self-Destruct
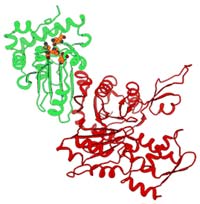
This ribbon diagram is a theoretical representation of the structure of adenovirus protease (red) bound to the cytoskeleton protein actin (green). The blue, green, and yellow balls show the location of the active site of the adenovirus protease. <br>
Scientists at the U.S. Department of Energy’s Brookhaven National Laboratory and their collaborators have discovered that some viruses can use the most abundant protein in the cells they are infecting to destroy the cells and allow new viruses to escape to infect others. The findings, described in the November 29, 2002, issue of the Journal of Biological Chemistry, build upon earlier Brookhaven research on how virus particles become infectious (see related story) and may lead to the design of more effective antiviral remedies.
“This is a new and philosophically interesting way for a virus to escape from cells,” said Brookhaven biologist Walter Mangel, a coauthor on the paper. “In essence, a protein in the infected cells can serve as the seed of the cells’ own destruction.”
Mangel’s group has previously shown that adenovirus — a virus that causes respiratory and gastrointestinal infections and also conjunctivitis — produces a protein-cleaving enzyme, or protease, to complete the maturation of newly synthesized virus particles. Similar to the way supportive scaffolding is removed after the completion of a construction project, this protease cleaves, or cuts out, viral “construction” proteins, leaving infectious virus particles behind.
This viral protease is produced in the cytoplasm in an inactive form, and must migrate to the nucleus to become activated in newly synthesized viral particles by two viral cofactors. Once activated, it can cleave several viral proteins to complete the viral maturation process. There were no indications that the protease could be activated in the cell’s cytoplasm.
When Mangel presented this research at a seminar at Princeton University, Clarence Schutt, a Princeton chemistry professor, pointed out that the amino acid sequence of one of the viral cofactors was dramatically similar to the sequence of actin, a cytoplasmic protein that gives shape and structure to cells. Mangel wondered if actin could activate the protease, and took some from Schutt’s lab back to Brookhaven to do the experiment.
The result: Incubating actin and the adenovirus protease increased the cleaving ability of the protease, just like the viral cofactor did, allowing the actin-protease complex to cleave actin itself, as well as other cellular skeleton proteins.
“When actin and other cytoskeleton proteins are destroyed,” Mangel explained, “a cell loses its shape and eventually breaks open, allowing the newly synthesized virus particles to escape and infect other cells.”
While other viruses are known to cleave actin and other cytoskeleton proteins as a means of breaking free to continue infection, none of them is known to use actin as a cofactor in this process, Mangel said. “Thus, the really interesting finding in this case is that actin is a cofactor for its own destruction,” Mangel said.
The next step for Mangel’s group will be to crystallize complexes of actin bound to the adenovirus protease to determine its atomic structure at the National Synchrotron Light Source at Brookhaven.
“The structure would then be used to find drugs to prevent the interaction between actin and the adenovirus protease,” Mangel said. “Such drugs could serve as a new type of antiviral agent.”
This work was done in collaboration with Mark Brown, Kevin McBride, Mary Lynn Baniecki, and Nancy Reich of Stony Brook University and Gerard Marriott of the University of Wisconsin. It was funded by the U.S. Department of Energy, which supports basic research in a variety of scientific fields, and the National Institutes of Health.
Media Contact
More Information:
http://www.bnl.gov/bnlweb/pubaf/pr/2002/bnlpr111902.htmAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















