High-performance computing reveals missing genes

Using an ephemeral supercomputer made up of computers from across the world, the mpiBLAST computational tool used by the researchers took only 12 hours instead of the 90 years it would have required if the work were performed on a standard personal computer.
The new study, reported in the journal BMC Bioinformatics, is the first large-scale attempt to identify undetected genes of microbes in the burgeoning GenBank DNA sequence repository that contains over 100 billion bases of DNA sequence. The genes uncovered may have important functions in the cell, but those functions need to be established by further experiment.
Skip Garner, executive director of VBI and professor of biological sciences at Virginia Tech, commented, “This is a perfect storm, where an overwhelming amount of data is analyzed by state-of-the-art computational approaches, yielding important new information about genes. These genes may be tomorrow's new targets for pharmaceutical research, for example to find new antibiotics or vaccines, which is extremely important since we need novel approaches to combat the emergence of new drug-resistant bugs.”
In the past few years, enormous progress has been made in sequencing technologies that allow scientists to produce astonishing amounts of sequence data. Today more than 1200 genome sequences of microbes are housed in the GenBank database. By far one of the biggest problems facing scientists is not generating the sequence data but reliably locating and assigning a function to the many genes in a genome, a process that scientists refer to as annotation. This process crucially depends on sophisticated computational tools. The field of bioinformatics is considered by many experts to have been started to address this very need.
João Setubal, associate professor at the Virginia Bioinformatics Institute and the Department of Computer Science at Virginia Tech, commented: “Scientists have known for a long time that publicly available databases of genomes have inconsistencies, errors, and gaps. Some genes are labeled with the wrong function and for others the function is unknown. But nobody had done a systematic study to verify how many genes were simply undetected. This is what we did in our study – discover the number of microbial genes that are under the radar.”
Scientists have developed different computer tools to help them in their efforts to locate and identify genes. Most of these tools work by building a model based on the features of the sequence and working out the likelihood that an individual segment codes for a gene. Comparing DNA segments with known gene sequences stored in GenBank complements this work. If a DNA segment is similar to the sequence of known genes, then the segment is likely to be a coding gene with a similar function.
Said Setubal, “Such approaches will not find genes that have unusual sequence properties. Furthermore they will not find those genes that have not been detected up to now and hence are not present in GenBank. Our results clearly show that there are many small protein-encoding genes in the genomes of microbes that have been systematically missed.”
The lowest estimate in the study placed the number of families of missing genes at 380 in the 780 genomes that were investigated. Said Setubal, “This number is most likely an underestimate since we have been conservative for the criteria we have used for finding these missing gene families.”
Wu Feng, associate professor in the Department of Computer Science and the Department of Electrical and Computer Engineering at Virginia Tech, remarked: “To facilitate the rapid discovery of missing genes in genomes, we used our mpiBLAST sequence-search tool to perform an all-to-all sequence search of the 780 microbial genomes that we investigated. This process entailed running on the order of tens of trillions of sequence searches with mpiBLAST. The all-to-all sequence search was done on an ephemeral supercomputer that aggregated more than 12,000 processor cores across seven different supercomputers, distributed across the United States. It reduced the search time from nearly 90 years, when computed on a personal computer, down to a mere 12 hours.”
Andrew Warren, a graduate assistant at VBI who has been working on this project as part of his PhD thesis, remarked: “At the outset of this project, the challenge was to create a method based on high-performance computing that could make meaningful predictions from such a large dataset. Through this work we were able to identify potential targets for future research and experimentation that can determine if these genes exist in vivo.”
Some of the preliminary work that is described in the current paper, specifically the computational and data management, was the winner of a distinguished paper award at the 2008 International Supercomputing Conference. The paper “Distributed I/O with ParaMEDIC: Experiences with a Worldwide Supercomputer,” recounted the computing experiences of an international team in finding missing genes in genomes and in constructing a genome similarity tree from the International Storage Challenge at the 2007 ACM/IEEE SC: The International Conference for High Performance Computing, Networking, Storage and Analysis.
Read the current paper:
Warren AS, Archuleta J, Feng W, Setubal JC (2010) Missing genes in the annotation of prokaryotic genomes. BMC Bioinformatics 11: 131. [PMID: 20230630]
Available on-line at http://www.biomedcentral.com/1471-2105/11/131
About VBI
The Virginia Bioinformatics Institute (http://www.vbi.vt.edu) at Virginia Tech is a premier bioinformatics, computational biology, and systems biology research facility that uses transdisciplinary approaches to science combining information technology, biology, and medicine. These approaches are used to interpret and apply vast amounts of biological data generated from basic research to some of today's key challenges in the biomedical, environmental, and agricultural sciences. With more than 240 highly trained multidisciplinary, international personnel, research at the institute involves collaboration in diverse disciplines such as mathematics, computer science, biology, plant pathology, biochemistry, systems biology, statistics, economics, synthetic biology, and medicine. The large amounts of data generated by this approach are analyzed and interpreted to create new knowledge that is disseminated to the world's scientific, governmental, and wider communities.
Media Contact
More Information:
http://www.vbi.vt.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
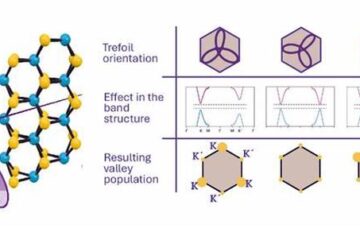
A novel universal light-based technique
…to control valley polarization in bulk materials. An international team of researchers reports in Nature a new method that achieves valley polarization in centrosymmetric bulk materials in a non-material-specific way…

How evolution has optimised the magnetic sensor in birds
The magnetic sense of migratory birds is probably based on the protein cryptochrome 4, and a genetic study has now provided further support for this theory. A team of researchers…
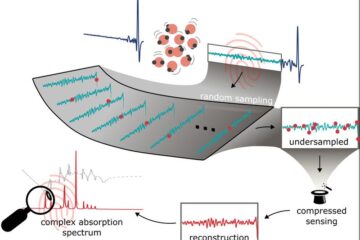
Molecular Fingerprint Beyond the Nyquist Frequency
Ultrafast laser spectroscopy allows the ascertainment of dynamics over extremely short time scales, making it a very useful tool in many scientific and industrial applications. A major disadvantage is the…





















