Carnivorous Plant Packs Big Wonders Into Tiny Genome
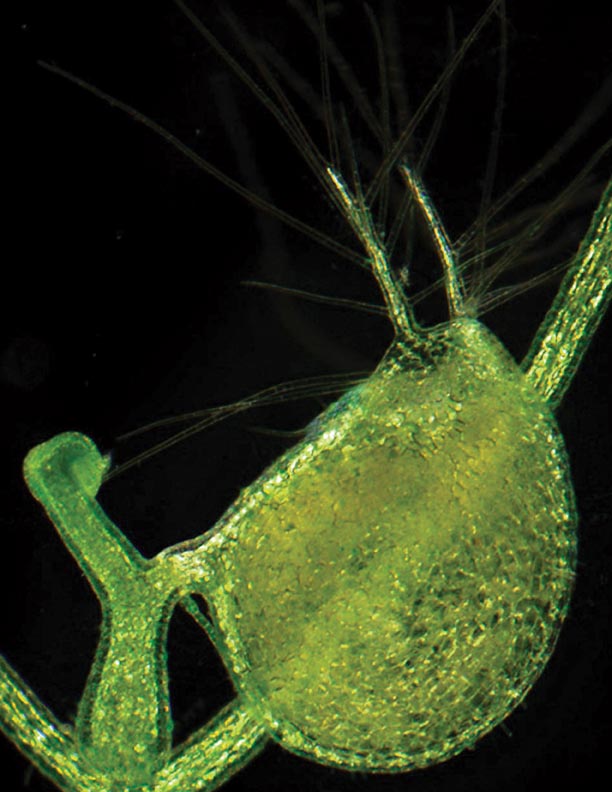
Credit: Enrique Ibarra-Laclette and Claudia Anahí Pérez-Torres. Light micrograph of the bladder of the carnivorous bladderwort plant, Utricularia gibba. A new study finds that this marvelous plant houses more genes than several well-known species, such as grape, coffee or papaya — despite having a much smaller genome.
Great, wonderful, wacky things can come in small genomic packages.
That’s one lesson to be learned from the carnivorous bladderwort, a plant whose tiny genome turns out to be a jewel box full of evolutionary treasures.
Called Utricularia gibba by scientists, the bladderwort is a marvel of nature. It lives in an aquatic environment. It has no recognizable roots. It boasts floating, thread-like branches, along with miniature traps that use vacuum pressure to capture prey.
A new study in the scientific journal Molecular Biology and Evolution breaks down the plant’s genetic makeup, and finds a fascinating story.
According to the research, the bladderwort houses more genes than several well-known plant species, such as grape, coffee or papaya — despite having a much smaller genome.
This incredibly compact architecture results from a history of “rampant” DNA deletion in which the plant added and then eliminated genetic material at a very fast pace, says University at Buffalo Professor of Biological Sciences Victor Albert, who led the study.
“The story is that we can see that throughout its history, the bladderwort has habitually gained and shed oodles of DNA,” he says.
“With a shrunken genome,” he adds, “we might expect to see what I would call a minimal DNA complement: a plant that has relatively few genes — only the ones needed to make a simple plant. But that’s not what we see.”
A unique and elaborate genetic architecture
In contrast to the minimalist plant theory, Albert and his colleagues found that U. gibba has more genes than some plants with larger genomes, including grape, as already noted, and Arabidopsis, a commonly studied flower.
A comparison with the grape genome shows U. gibba’s genetic opulence clearly: The bladderwort genome, holding roughly 80 million base pairs of DNA, is six times smaller than the grape’s. And yet, the bladderwort is the species that has more genes: some 28,500 of them, compared to about 26,300 for the grape.
U. gibba is particularly rich in genes that may facilitate carnivory — specifically, those that enable the plant to create enzymes similar to papain, which helps break down meat fibers. The bladderwort is also rich in genes linked to the biosynthesis of cell walls, an important task for aquatic species that must keep water at bay.
“When you have the kind of rampant DNA deletion that we see in the bladderwort, genes that are less important or redundant are easily lost,” Albert says. “The genes that remain — and their functions — are the ones that were able to withstand this deletion pressure, so the selective advantage of having these genes must be pretty high.
“Accordingly, we found a number of genetic enhancements, like the meat-dissolving enzymes, that make Utricularia distinct from other species.”
Much of the DNA the bladderwort deleted over time was noncoding “junk DNA” that contains no genes, Albert says.
High gene turnover
The study included partners from UB, the Universitat de Barcelona in Spain, the Laboratorio Nacional de Genómica para la Biodiversidad (LANGEBIO) in Mexico and the Instituto de Ecología in Mexico.
To determine how the bladderwort evolved its current genetic structure, the team compared the plant to four related species. What they uncovered was a pattern of rapid DNA alteration.
As Albert explains, “When you look at the bladderwort’s history, it’s shedding genes all the time, but it’s also gaining them at an appreciable enough rate, permitting it to stay alive and produce appropriate adaptations for its unique environmental niche.”
In the realm of DNA gain, the study found that U. gibba has undergone three duplication events in which its entire genome was replicated, giving it redundant copies of every gene.
This fast-paced gene gain was balanced out by swift deletion. Evidence for this phenomenon comes from the fact that the plant has a tiny genome despite its history of genetic duplication. In addition, the plant houses a high percentage of genes that don’t have close relatives within the genome, which suggests the plant quickly deleted redundant DNA acquired through duplication events.
The study was supported by the National Science Foundation. It builds on the work of Albert and other team members, who reported in the journal Nature in 2013 that the bladderwort’s genome was comprised almost entirely of useful, functional genes and their controlling elements, in contrast to species like humans, whose genomes are more than 90 percent “junk DNA.”
Contact Information
Contact: Charlotte Hsu, chsu22@buffalo.edu
University at Buffalo
716-645-4655
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

A universal framework for spatial biology
SpatialData is a freely accessible tool to unify and integrate data from different omics technologies accounting for spatial information, which can provide holistic insights into health and disease. Biological processes…

How complex biological processes arise
A $20 million grant from the U.S. National Science Foundation (NSF) will support the establishment and operation of the National Synthesis Center for Emergence in the Molecular and Cellular Sciences (NCEMS) at…

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…





















