Study of transplanted hearts reveals risk gene for cardiovascular disease
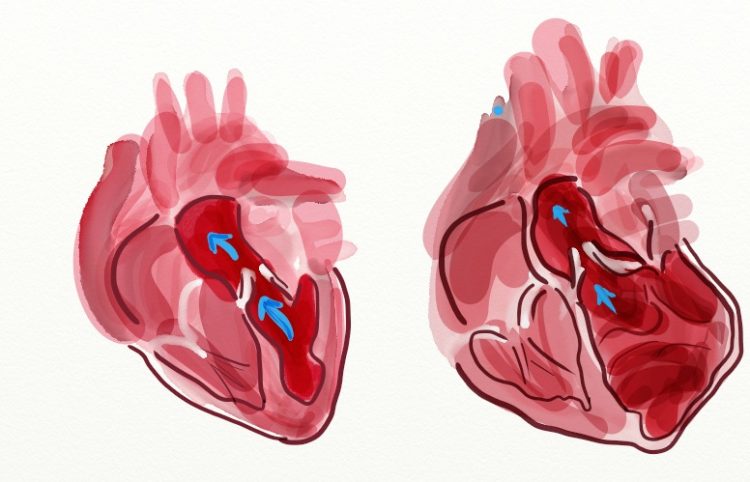
While a normal heart pumps a certain amount of blood (left), the dilated heart has much lower pumping capacity (right) Illustration: Dr. Eleonora Adami, MDC
We know of many genes with variants that make us particularly prone to cardiovascular disease. But there are gaps in our knowledge. It is not just the genes themselves but also the way they are expressed that influences the risk of disease.
In some cases, DNA regions that control how genetic information is copied onto RNA molecules are changed. The amount of RNA and whether this temporary information carrier is subsequently modified influence the development of disease.
To investigate these processes at RNA-level, however, there is a serious lack of research material. “It is extremely hard to get human heart tissue for genetic studies,” says Professor Norbert Hübner, one of the leading researchers involved in the study. “That’s why we still don’t know all the variants of risk genes by a long chalk.”
Together with a team from the Helmholtz Zentrum München and researchers from the Netherlands and Singapore, the group leader at the Max Delbrück Center for Molecular Medicine (MDC), the Charité – Universitätsmedizin Berlin and the Deutsches Zentrum für Herz-Kreislauf-Erkrankungen (DZHK) has just published a paper in the journal Genome Biology. It is the largest genetic study yet undertaken comparing the entire transcriptome of heart tissue in healthy and diseased individuals.
Heart transplants facilitate large genetic study
The research team was keen to discover which genes increase the risk of dilated cardiomyopathy (DCM), an inherited myocardial insufficiency that can lead to heart failure and sudden death.
During heart transplants, the scientists collected biopsies from the left ventricle of 97 patients with DCM and 108 healthy donors. They analysed the transcriptome and managed to identify 228 genes which are expressed differently in DCM patients and healthy subjects.
Certain RNA molecules or RNA modifications also occured with varying frequency. The research team confirmed that these regulatory differences are found, above all, in known DCM risk genes. But they also identified sixty new genes that are active in the heart and have not so far been linked to DCM.
Early detection of individuals at risk
According to Dr Matthias Heinig, lead author and group leader at the Helmholtz Zentrum München, “Our data are a valuable resource for cardiovascular research as a whole.” They make it easier to evaluate potential risk genes and offer researchers new indications for developing drugs and diagnostic tests.
“A test of this kind should identify people with an increased risk at an early stage,” says Heinig. “Then they can be treated in good time or change their lifestyle as a precautionary measure.”
“Although there are already plenty of clinical indications for many cardiovascular diseases,” Hübner, the physician, adds, “we hope a transcriptome analysis will be able to improve these predictions.”
In Hübner’s opinion the study is a major step forward for another reason, too: “We now understand DCM better. Our study has discovered adjusting screws for the development of the disease that go well beyond classic gene mutations.”
Matthias Heinig et al. (2017): „Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy.“ Genome Biology. doi:10.1186/s13059-017-1286-z (Open Access)
https://www.mdc-berlin.de/1150679/ – Website of the group “Genetics and Genomics of Cardiovascular Diseases”
Media Contact
More Information:
http://www.mdc-berlin.de/All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

A universal framework for spatial biology
SpatialData is a freely accessible tool to unify and integrate data from different omics technologies accounting for spatial information, which can provide holistic insights into health and disease. Biological processes…

How complex biological processes arise
A $20 million grant from the U.S. National Science Foundation (NSF) will support the establishment and operation of the National Synthesis Center for Emergence in the Molecular and Cellular Sciences (NCEMS) at…

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…





















