Numbers count in the genetics of moles and melanomas
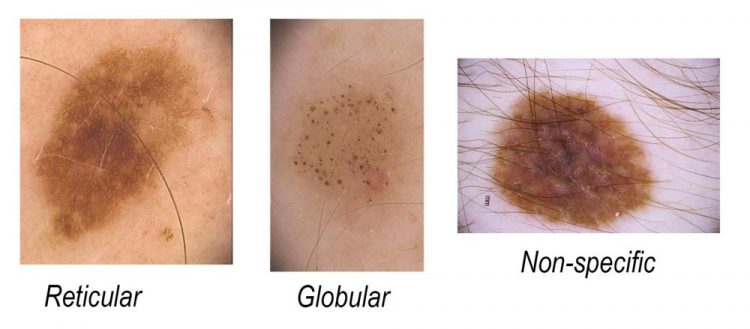
Three key mole classes, reticular, globular and non-specific were magnified under a dermoscope to assess their pattern and risk factors. Credit: The University of Queensland
University of Queensland scientists have identified a way to help dermatologists determine a patient's risk of developing melanoma.
UQ Diamantina Institute researcher Associate Professor Rick Sturm said the team uncovered the specific gene variations affecting the number and types of moles on the body and their role in causing skin cancer.
“The goal was to investigate the genetic underpinnings of different mole classes or 'naevi types' and understand how these affect melanoma risk,” Dr Sturm said.
“Based on our work, the number of moles in each category can give a more complete assessment of melanoma risk rather than just the number of moles alone.”
Three key mole classes, reticular, globular and non-specific were magnified under a dermoscope to assess their pattern and risk factors.
“We found people who had more non-specific mole patterns increased their melanoma risk by two per cent with every extra mole carried,” he said.
“As we age, we tend to increase the amount of non-specific moles on our body, and the risk of developing melanoma increases.”
Dr Sturm said globular and reticular mole patterns were also found to change over time.
“Globular patterns were shown to decrease as we get older, typically petering out after the age of 50 to 60,” he said.
“Reticular moles also decreased over time but were likely to head down a more dangerous path and develop into the non-specific pattern.”
A cohort of more than 1200 people, half melanoma patients, were recruited into the almost nine-year study.
Their results were then overlayed with genetic testing, which found variations in four major genes.
“We found some major relationships between genes and the number of moles and patterns when looking at the DNA,” Dr Sturm said.
“Certain gene types influenced the number of different naevi types — for example, the IRF4 gene was found to strongly influence the number of globular naevi found on the body.”
The findings will help dermatologists to better understand mole patterns and provide more holistic care to patients who may be at risk of melanoma.
“For a long time, clinicians have been interested in how pigmented moles relate to melanoma and melanoma risk,” he said.
“With the availability of dermoscopes and imaging, these results provide a new layer of understanding to guide clinical practice.”
###
This research is published in Journal of Investigative Dermatology (DOI: 10.1016/j.jid.2019.05.032)
Media: Associate Professor Rick Sturm, r.sturm@ uq.edu.au; Faculty of Medicine Communications, med.media@uq.edu.au, +61 7 3365 5118, +61 436 368 746.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















