Tailor-made Cancer Drugs: Wave of the Future?

Washington University chemist offers radical new strategy in fight against cancer
Today, even the best cancer treatments kill about as many healthy cells as they do cancer cells but John-Stephen A. Taylor, Ph.D., professor of chemistry at Washington University in St. Louis, has a plan to improve that ratio. Over the last several years, Taylor has begun to lay the conceptual and experimental groundwork for a radical new strategy for chemotherapy — one that turns existing drugs into medicinal “smart bombs,” if you will.
All DNA is formed of three basic components: a phosphate and a sugar, which combine to form the sides of the double helix “ladder,” and a base that forms the ladder’s “rungs.” All variances in DNA, including cancerous mutations, are the result of unique sequencing of the four types of bases, denoted A, G, C and T.
Taylor’s approach, described as “nucleic acid-triggered catalytic drug release,” is essentially a sophisticated drug releasing system, one that is able to recognize and use cancerous sequences as triggering mechanisms for the very drugs that fight them.
“The beauty of this system is that it could use already-approved FDA drugs,” Taylor explained. “So all I have to worry about is getting FDA approval on the general releasing mechanism, and then I can incorporate whatever anticancer drugs are currently on the market.”
Taylor discussed his work at the 40th annual New Horizons in Science Briefing, a function of the Council for the Advancement of Science Writing. He spoke Oct. 27, 2002 at Washington University in St. Louis, which hosted the event.
Guiding drugs to their ’parking spot’
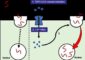
In nucleic acids, Nature has already determined the rules of base pairing — A binds with T and G pairs with C — a system called “Watson-Crick base-pairing,” named for the discoverers of the double helix. Recent advances in biotechnology have given doctors the ability to profile a patient’s genetic information, taken during a biopsy, using something called a DNA chip, which can identify unique or uniquely overexpressed messenger RNA (mRNA). Messenger RNA is a single-stranded RNA molecule that encodes information to make a protein, using the same bases as DNA except that U replaces T. Taylor’s idea is to employ this information as a genetic roadmap, guiding drug components to where they should “park” amongst the millions of base pair “spaces.”
Taylor’s system is built on three components: a “prodrug,” or a dormant form of a drug; a catalyst that activates the prodrug; and a nucleic acid triggering sequence, designed to match and interlock with a unique or uniquely overexpressed strand of RNA in cancerous cells. The RNA binding drug components will be fashioned out of Peptide Nucleic Acid (PNA), which is identical to DNA, but replaces the sugar backbone with a “peptide” or protein backbone. The benefit is that a single strand of RNA actually binds tighter to a strand of PNA than it does to itself.
So, the prodrug and the catalytic components each contain a PNA strand that is complementary to the cancer cell’s mRNA, allowing them to bind right next to one another in the cancer cell. This close proximity enables a chemical reaction to occur between them, resulting in the release of a cytotoxic drug which kills the cancer cell. Although the medication might encounter healthy cells in its travels, it would not harm them because the RNA triggering sequence would not be present, or else present in a much lower amount, and the drug could not be released.
This new “rational” design doesn’t stop there — it could be the answer to all sorts of viral diseases such as AIDS, hepatitis and herpes, and could even help guard against new biologically engineered viruses that we haven’t yet imagined.
“Here’s my vision of the future,” Taylor said. “You go to a doctor’s office and take a biopsy, which is then run through a DNA chip analysis machine allowing the appropriated triggering sequence to be identified. This information is then passed to an automated synthesis machine and, iIdeally, the catalytic and prodrug components can be synthesized and administered to you within hours.”
In related work, Taylor said he will be using overexpressed RNA sequences to help target drugs in research with Washington University colleague Karen Wooley, Ph.D., associate professor of chemistry, and other collaborators. The group hopes to splice Taylor’s RNA-docking molecules to Wooley’s new breed of nanoparticles for on-the-mark, stay-put delivery of diagnostic and disease-fighting agents.
Questions
Contact: Tony Fitzpatrick, senior science editor, Washington University in St. Louis, (314) 935-5272; tony_fitzpatrick@aismail.wustl.edu
Media Contact
All latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…