Study finds vulnerability in malaria parasite
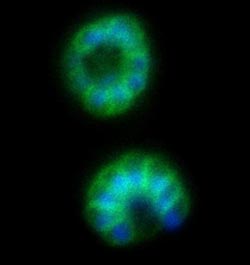
These are malaria parasites (labeled with flourescent protein) in the late stages of development, superimposed on a field of red blood cells. The many nuclei of the parasites' daughter cells are labeled in blue, and the plasma membranes surrounding the daughter cells are labeled in green. Imidazopyrazine treatment disrupts formation of the membranes around the daughter cells.<br><br>Credit: Marcus C.S. Lee, Ph.D./Columbia University Medical Center<br>
An international team of scientists, including researchers at Columbia University Medical Center (CUMC), has identified a key metabolic enzyme that common malaria parasites require for survival at each stage of infection in humans.
The findings raise the possibility of a new approach to combating malaria, one of the world's deadliest diseases. The study was published today in the online edition of the journal Nature.
“Perhaps the most exciting aspect of our findings is that this enzyme is required at all stages of the parasites' life cycle in humans,” said co-first author Marcus C.S. Lee, PhD, associate research scientist in microbiology & immunology at CUMC.
“This is important because most antimalarials are effective at killing the parasites only as they circulate in the bloodstream. However, the parasites can hide in the liver for years before reemerging and triggering a relapse of the disease. By identifying this enzyme, we may be able to develop a new way to kill the parasites in their dormant stage.”
The other co-first author is Case W. McNamara, PhD, research investigator at the Genomics Institute for the Novartis Research Foundation. The study leaders are Elizabeth A. Winzeler, PhD, professor of pharmacology and drug discovery at University of California San Diego, and Thierry Diagana, head of Novartis Institute for Tropical Diseases in Singapore.
The enzyme — phosphatidylinositol 4-kinase (PI4K) — was found by screening more than a million drug compounds against Plasmodium falciparum, the parasite responsible for the most lethal form of malaria. Using this screen, the researchers found a class of compounds known as imidazopyrazines, which are capable of killing several species of Plasmodium at each stage of the parasites' life cycle in its vertebrate host. Also important, the compounds had no effect on human cells.
The researchers identified the target of the imidazopyrazines by evolving parasite cell lines that were resistant against the drugs and then analyzing the parasites' genomes for the changes responsible for conferring resistance. Those genetic changes pointed to the gene that encodes PI4K.
The CUMC team, led by David Fidock, PhD, professor of microbiology & immunology and medical sciences (in medicine), used novel genetic tools to confirm that PI4K was being directly targeted by the imidazopyrazines.
Then, using cellular imaging, the CUMC team found that imidazopyrazines interfere with the function of PI4K on the parasite Golgi (the organelle that packages proteins for delivery to other cellular destinations). “We think that disrupting the function of PI4K at the Golgi stops the parasite from making new membranes around its daughter cells, thereby preventing the organism from reproducing,” said Dr. Lee.
Because PI4K is also found in humans, Dr. Winzeler said, the next challenge is to develop a drug that retains selectivity between the parasite and human versions of the enzyme. “As we now know the identity of this protein and hope to soon solve its structure, this task should be much easier,” she said.
The paper is titled, “Targeting Plasmodium phosphatidylinositol 4-kinase to eliminate malaria.” The other contributors are: Chek Shik Lim (Novartis Institutes for Tropical Disease, Singapore), Siau Hoi Lim (Novartis), Jason Roland (UCSD), Advait Nagle (UCSD), Oliver Simon (Novartis), Bryan K.S. Yeung (Novartis), Arnab K. Chatterjee (UCSD), Susan L. McCormack (UCSD), Micah J. Manary (UCSD), Anne-Marie Zeeman (Biomedical Primate Research Centre, Rijswijk, the Netherlands), Koen J. Dechering (TropIQ Health Sciences, Nijmegen, the Netherlands), T.R. Santha Kumar (CUMC), Philipp P. Henrich (CUMC), Kerstin Gagaring (UCSD), Maureen Ibanez (UCSD), Nobutaka Kato (UCSD), Kelli L. Kuhen (UCSD), Christoph Fischli (Swiss Tropical and Public Health Institute, Basel, Switzerland), Matthias Rottmann (Swiss Tropical and Public Health Institute and University of Basel, Basel, Switzerland), David M. Plouffe (UCSD), Badry Bursulaya (UCSD), Stephan Meister UCSD), Lucia Rameh (Boston University, Boston), Joerg Trappe (Novartis Institutes for BioMedical Research, Basel, Switzerland), Dorothea Haasen (Novartis, Basel, Switzerland), Martijn Timmerman (TropIQ Health Sciences), Robert W. Sauerwein (Trop IQ Health Sciences and Radboud University, Nijmegen Medical Centre, Nijmegen, the Netherlands), Rossarin Suwanarusk (Agency for Science Technology and Research, Biopolis, Singapore), Bruce Russell (Agency for Science Technology and Research and National University of Singapore, National University Health System, Singapore), Laurent Renia (Agency for Science Technology and Research), Francois Nosten (University of Oxford, Oxford, UK, and Mahidol University, Mae Sot, Thailand), David C. Tully (UCSD), Clemens HM Kocken (Biomedical Primate Research Centre), Richard J. Glynne (UCSD), Christophe Bodenreider (Novartis, Singapore), and Thierry T. Diagana (Norvartis, Singapore).
The authors wish to disclose the following, which may be considered a conflict of interest: C.W.M, C.S.L, S.H.L, J.R., O.S., B.K.S.Y., K.L.K., K.G., D.M.P., B.B., J.T., D.H., D.T., R.J.G., C.B. and T.T.D. are employed by Novartis. C.W.M, J.R., K.L.K., B.B., J.T., D.H., D.T., R.J.G, T.T.D. and E.A.W. own shares of Novartis AG stock. E.A.W. has received grants from Novartis.
The study was supported by grants from the Wellcome Trust (WT078285 and WT096157) and funding from the Medicines for Malaria Venture at the Genomics Institute of the Novartis Research Foundation, the Swiss Tropical and Public Health Institute, Columbia University, the Novartis Institute for Tropical Diseases, the Singapore Immunology Network and Horizontal Programme on Infectious Diseases under the Agency Science Technology and Research, and the Wellcome Trust (UK). SMRU is sponsored by the Wellcome Trust of Great Britain, as part of the Oxford Tropical Medicine Research Programme of Wellcome Trust-Mahidol University. E.A.W. and D.A.F. are supported by grants from the Bill and Melinda Gates Foundation, MMV, and the NIH (R01AI090141 to E.A.W. and R01085584 and R01079709 to D.A.F.).
Columbia University Medical Center provides international leadership in basic, preclinical, and clinical research; medical and health sciences education; and patient care. The medical center trains future leaders and includes the dedicated work of many physicians, scientists, public health professionals, dentists, and nurses at the College of Physicians and Surgeons, the Mailman School of Public Health, the College of Dental Medicine, the School of Nursing, the biomedical departments of the Graduate School of Arts and Sciences, and allied research centers and institutions. Columbia University Medical Center is home to the largest medical research enterprise in New York City and State and one of the largest faculty medical practices in the Northeast. For more information, visit cumc.columbia.edu or columbiadoctors.org.
Media Contact
More Information:
http://www.columbia.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















