Basic RNA enzyme research promises single-molecule biosensors
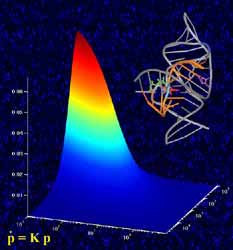
Studying RNA enzymes. The graph in the foreground shows how the enzyme’s catalytic activity is related to the rates at which the molecule folds and unfolds. These rates were measured by single-molecule fluorescence microscopy, where individual molecules light up as bright spots shown in the background. Also depicted, top right, is a ribbon-and-stick representation of the crystal structure of the folded RNA enzyme. <br>
Research aimed at teasing apart the workings of RNA enzymes eventually may lead to ways of monitoring fat metabolism and might even assist in the search for signs of life on Mars, according to University of Michigan researcher Nils Walter. His latest work was published online in the Proceedings of the National Academy of Sciences June 24.
Walter and associates at U-M and colleague Xiaowei Zhuang and associates at Harvard University, use techniques that allow them to study single molecules of RNA enzymes, also known as ribozymes. Like the more familiar protein enzymes, RNA enzymes accelerate chemical reactions inside cells. Researchers want to learn how changes in ribozyme molecules affect their activity, both to better understand how evolution has shaped ribozymes to carry out their duties and to find ways of manipulating them for useful purposes.
In the recent research, Walter’s group combined a technique called single-molecule fluorescence resonance energy transfer (FRET) with mathematical simulations to study a ribozyme involved in the replication of a tobacco-infecting virus. Just as a protein enzyme is not a static structure, a ribozyme also changes shape, cycling back and forth between its compact, catalytically active form and its inactive, extended form. Single-molecule FRET allowed the researchers to directly observe and measure how quickly the ribozyme switched forms and how these rates changed when various parts of the molecule were altered.
With the addition of mathematical simulations, the researchers also could investigate how changing parts of the ribozyme molecule affected its ability to catalyze chemical reactions. They were surprised to find that modifications they made anywhere on the molecule—even far from the site where the chemical reaction occurs—affected the rate of catalysis.
That’s much like what is known to happen in protein enzymes, but until now there was no evidence that ribozymes behaved similarly, said Walter, a Dow Corning Assistant Professor of Chemistry.
“It’s been known for a couple of years now that if you modify something on a protein enzyme that you think is pretty far away from the catalytic core—where the chemistry is actually happening—you see that the chemistry is affected directly,” Walter said. “This has led to the idea that there is a network of motions that make a protein enzyme act as a whole. We are proposing for the first time that this also happens with RNA enzymes.”
Getting a grasp on how ribozymes work is important for answering fundamental questions of biology, Walter said, but the work may also lead to practical applications. In particular, Walter and U-M collaborators Robert T. Kennedy, the Hobart H. Willard Professor of Chemistry and Pharmacology, and Jens-Christian Meiners, assistant professor of physics and assistant research scientist, Biophysics Research Division, are exploring their use as biosensors. The idea is to selectively turn on a ribozyme molecule that catalyzes a reaction to generate a product that gives off a specific fluorescent signal only when a particular type of molecule binds.
“When you can do that on the single-molecule level, as we can do now, then you have the smallest possible biosensor,” Walter said. Such sensors could be designed to detect important hormones like leptin, which is involved in fat metabolism. With such a tool, “you could detect how a single cell makes leptin and ask how much the cell makes when the environment changes,” he said.
In another project, funded by NASA, the researchers hope to develop a biosensor that could be sent to Mars to snoop around for amino acids or other signs that life might once have existed on the planet.
“These projects are still in the development stage,” Walter said. “But the technology we are developing here to ask some fundamental biological questions will ultimately help us learn how to design biological sensors with many potential applications.”
Media Contact
More Information:
http://www.umich.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















