DNA separation by entropic force offers better resolution
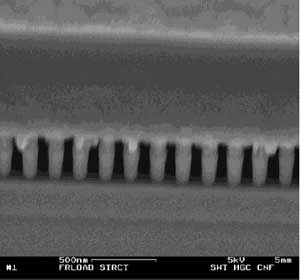
Scanning electron micrograph of a cross section of the finished device. An array of densely spaced nanopillars constitutes the entropically unfavorable region. The pillar spacing was 135 nm and their width approximately 80 nm. Copyright © Cornell
Cornell University researchers have demonstrated a novel method of separating DNA molecules by length. The technique might eventually be used to create chips or other microscopic devices to automate and speed up gene sequencing and DNA fingerprinting.
The method, which uses a previously discovered entropic recoil force, has better resolution — that is, better ability to distinguish different lengths — than others tried so far, the researchers say. They separated DNA strands of two different lengths, using their own nanofabricated device, and demonstrated that modifications would make it possible to separate strands of many different lengths.
A description of the experiment is scheduled to be published in the Oct. 1, 2002, issue of the journal Analytical Chemistry by graduate student Mario Cabodi, postdoctoral researcher Stephen Turner and Harold Craighead, the C.W. Lake Jr. Professor of Productivity.
The traditional method of separating DNA is gel electrophoresis, in which a strand is cut into many pieces and passed through a porous gel, where shorter lengths will move faster and farther than longer ones. From the distribution of the fragments, information about the genetic content can be determined. Researchers at Cornell and elsewhere have been experimenting with a variety of devices that replace the porous gel with microscopic sieves made by the same techniques used to manufacture electronic circuits.
Previously, Turner, Cabodi and Craighead studied the physics governing the movement of molecules through these sieves. Ordinarily, a long chain DNA molecule in liquid will clump into a roughly spherical shape, and to move through a sieve it must uncoil and slide in lengthwise. The researchers found that this movement involves an entropic force which causes DNA molecules that are only partially within a sieve to withdraw when the force pulling them in — an electric field — is removed. The effect is similar to a slippery chain falling from a table. If the chain is either entirely on the table or entirely on the floor, it will not move, but if part of it is hanging over the edge of the table it will eventually all spill onto the floor. In the case of DNA molecules in liquid, the effect results from the motion of segments in the chain molecule as they interact with the beginning of the barrier. The force is called “entropic” because the molecule moves out of the restricted space of the sieve into an open area where it can be more disordered.
In the latest experiment, the researchers used the Cornell Nanofabrication Facility (CNF) to make a sieve consisting of a forest of tiny pillars 80 nanometers wide, spaced about 135 nanometers apart, just large enough for a DNA molecule to pass through lengthwise. (A nanometer is a billionth of a meter, approximately three times the diameter of a silicon atom.) The researchers inserted a mix of DNA strands of two different lengths in a space next to the pillars and applied an electric field in a short pulse. The length of the pulse was adjusted so that shorter molecules were able to move completely into the pillar region, while longer ones could move only partway in. When the field was turned off, the longer strands recoiled, while the shorter ones, completely inside the pillar region, did not.
DNA molecules are too small to be observed directly, but for the experiment they were stained with a fluorescent dye, and the light given off was visible under a conventional microscope.
In further tests, the researchers showed that by applying a series of pulses of different durations the method could be used to separate strands of many different lengths. Another approach to separating strands of many lengths, they said in their paper, might be a “cascade” of similar separation devices. These methods would also improve the resolution, they said. They predicted that such a device could separate DNA molecules “in a particularly challenging length range with higher resolution than any other known method.”
The principle of entropic recoil, they said, also could be applied by using various types of porous membranes and ceramic filters, as well as nanofabricated sieves. The technique could also be applied to proteins and other polymers, they added.
The paper in Analytical Chemistry is titled “Entropic Recoil Separation of Long DNA Molecules.” The research was funded by the National Institutes of Health. CNF is supported by the National Science Foundation.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…

Simplified diagnosis of rare eye diseases
Uveitis experts provide an overview of an underestimated imaging technique. Uveitis is a rare inflammatory eye disease. Posterior and panuveitis in particular are associated with a poor prognosis and a…

Targeted use of enfortumab vedotin for the treatment of advanced urothelial carcinoma
New study identifies NECTIN4 amplification as a promising biomarker – Under the leadership of PD Dr. Niklas Klümper, Assistant Physician at the Department of Urology at the University Hospital Bonn…





















