Pinpointing Protein Locations
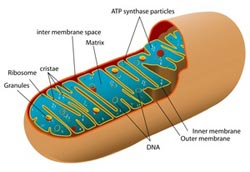
Diagram of a mitochondrion<br>
Scientist from MIT have now developed a technique that can tag all of the proteins in a particular region of a cell, allowing them to more accurately map those proteins.
“There was no previous high-quality map of the matrix subdomain of mitochondria, and now we have one” said Alice Ting, the Ellen Swallow Richards Associate Professor of Chemistry at MIT. “We’re still really far from that goal, but the overarching motivation is to get closer to that goal.”
This innovative technique combines the strengths of two existing techniques — microscopic imaging and mass spectrometry — to map proteins in a specific cell location and generate a comprehensive list of all the proteins in that area.
In a paper appearing in the Jan. 31 online edition of Science, Rhee and colleagues used the new technique to identify nearly 500 proteins located in the mitochondrial matrix — the innermost compartment found in mitochondria, which can be thought of as the power houses of the cell where energy is generated. Previous attempts to map the entire set of proteins in the matrix (proteome) yielded a list of only 37 proteins.
Protein labeling
Using fluorescence or electron microscopy, scientists can determine protein locations with high resolution, but only a handful of a cell’s ~20,000 proteins can be imaged at once. “It’s a bandwidth problem,” Ting says. “You certainly couldn’t image all the proteins in the proteome at once in a single cell, because there’s no way to spectrally separate that many channels of information.”
With mass spectrometry, which uses ionization to detect the mass and chemical structure of a compound, scientists can analyze a cell’s entire complement of proteins in a single experiment. However, the process requires dissolving the cell membrane to release a cell’s contents, which jumbles all of the proteins together. By purifying the mixture and extracting specific organelles, it is then possible to figure out which proteins were in those organelles, but the process is messy and often unreliable.
The new MIT approach tags proteins within living cells before mass spectrometry is done, allowing spatial information to be captured before the cell is broken apart. This information is then reconstructed during analysis by noting which proteins carry the location tag.
The new system makes use of a chemical tag that includes biotin, one of the B vitamins. To label proteins with biotin, the researchers first designed a new enzyme they dubbed APEX. This enzyme is a *peroxidase*, meaning that it removes an electron and a proton in a reaction known as oxidation.
“What you do is tag the proteins with biotin while the cell is still alive, and then you just pull out those proteins,” Ting says. “Therefore you bypass all of the problems that are associated with trying to purify regions of cells and organelles, because you don’t have to anymore.”
A comprehensive list
To demonstrate the technique’s power, the researchers created a comprehensive list of the proteins found in the mitochondrial matrix. Most of a cell’s energy generation takes place in mitochondria, as well as many biosynthetic processes.
Using the new method, the team increased the number of proteins known to be located in the mitochondrial matrix. “There was no previous high-quality map of the matrix subdomain of mitochondria, and now we have one,” says Ting, adding that this new wealth of information should help biologists to learn more about the functions of many of those proteins.
Already, the team has found that an enzyme called ppox — involved in synthesizing heme, the iron-porphyrin complex found in hemoglobin — is not located where biologists had thought it was. As heme precursors move through the biosynthetic pathway, they are shuttled to different parts of the cell. Finding that ppox is in the matrix means that there must be unknown transporter proteins bringing heme precursors into the matrix, Ting says.
The researchers are now investigating proteins found in another compartment of the mitochondria, the intermembrane space. They are also modifying the chemistry of the labeling system so they can use it for other tasks such as mapping the topology of membrane proteins and detecting specific protein-protein interactions.
The lead scientists of this research are Hyun-Woo Rhee (former MIT postdoc, currently Assistant Professor, School of Nano-Bioscience and Chemical Engineering, UNIST) and Peng Zou, who received a PhD from MIT in 2012.
REFERENCE:
Hyun-Woo Rhee, Peng Zou, Namrata D. Udeshi, Jeffrey D. Martell, Vamsi K. Mootha, Steven A. Carr, and Alice Y. Ting. 2013. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 1230593. Published online 31 January 2013 [DOI:10.1126/science.1230593]
Journal information
ScienceXpress
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

A universal framework for spatial biology
SpatialData is a freely accessible tool to unify and integrate data from different omics technologies accounting for spatial information, which can provide holistic insights into health and disease. Biological processes…

How complex biological processes arise
A $20 million grant from the U.S. National Science Foundation (NSF) will support the establishment and operation of the National Synthesis Center for Emergence in the Molecular and Cellular Sciences (NCEMS) at…

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…





















