"Sloppy Genes" Behave Like Their Neighbours

New Findings Reveal That The Regulation Of Gene Expression Is Much Less Strictly Controlled Than Was Previously Thought
The inaugural issue of Journal of Biology features groundbreaking research that challenges the traditional view of how genes are controlled. Our current understanding of gene expression, the fundamental process by which proteins are made from the instructions encoded in DNA, is that the process is tightly controlled so that the correct amount of each protein is produced in the right place at the right time. The new research, by Paul Spellman and Gerald Rubin of Howard Hughes Medical Institute and University of California Berkeley, indicates that some groups of around 15 genes that just happen to be located next to each other on chromosomes are instead routinely expressed together. Genes within a block are not related to one another by structure or function: they are simply neighbours within the genome. These conclusions were drawn from experiments using DNA-coated microarrays to monitor the expression of all the genes in the genome of the fruitfly.
Microarray experiments allow researchers to look at the expression of a large number of genes in a single experiment and ask which genes are being turned on (or expressed) in a particular cell type, at a certain time, under defined conditions. Spellman and Rubin analysed gene expression data from 88 independent microarray experiments using DNA extracted from the fruitfly, Drosophila melanogaster. Then instead of the usual practice of searching the microarray data for groups of genes that showed similar expression profiles, they arranged the expression data in the order in which the genes occur along the fly’s chromosomes.
To their surprise, Spellman and Rubin found over 200 large groups of adjacent and similarly expressed genes. Each of these groups contained between 10 and 30 genes, and together the groups accounted for approximately 20% of the genes in the fly’s genome. This suggests that there is a previously unsuspected mechanism of regulating of gene expression, which works on blocks of genes depending on their position along a chromosome.
Having ruled out other possible explanations, Spellman and Rubin speculate that the expression of gene groups might prove to reflect some feature of the chromatin – the packaging of DNA and proteins into chromosomes – that, in allowing an ‘important’ gene to be expressed, also leads to the expression of neighbouring genes, more by accident than design. This somewhat ‘sloppy’ regulation of gene expression may be used when it is relatively unimportant how much of each protein is produced, they suggest, whereas precise control of the gene is used when the level of a protein is critical.
Only time will tell if the expression of groups of genes located next to each other is a universal feature of higher organisms but Spellman and Rubin speculate that “as further experiments are carried out it may be found that our observation will grow to include all genes”.
Media Contact
More Information:
http://jbiol.comAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Why getting in touch with our ‘gerbil brain’ could help machines listen better
Macquarie University researchers have debunked a 75-year-old theory about how humans determine where sounds are coming from, and it could unlock the secret to creating a next generation of more…

Attosecond core-level spectroscopy reveals real-time molecular dynamics
Chemical reactions are complex mechanisms. Many different dynamical processes are involved, affecting both the electrons and the nucleus of the present atoms. Very often the strongly coupled electron and nuclear…
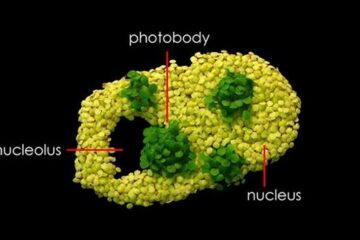
Free-forming organelles help plants adapt to climate change
Scientists uncover how plants “see” shades of light, temperature. Plants’ ability to sense light and temperature, and their ability to adapt to climate change, hinges on free-forming structures in their…