Scientists have waited months for access to highly accurate protein structure prediction since DeepMind presented remarkable progress in this area at the 2020 Critical Assessment of Structure Prediction, or CASP14, conference. The wait is now over.
Researchers at the Institute for Protein Design at the University of Washington School of Medicine in Seattle have largely recreated the performance achieved by DeepMind on this important task. These results will be published online by the journal Science on Thursday, July 15.
Unlike DeepMind, the UW Medicine team’s method, which they dubbed RoseTTAFold, is freely available. Scientists from around the world are now using it to build protein models to accelerate their own research. Since July, the program has been downloaded from GitHub by over 140 independent research teams.
Proteins consist of strings of amino acids that fold up into intricate microscopic shapes. These unique shapes in turn give rise to nearly every chemical process inside living organisms. By better understanding protein shapes, scientists can speed up the development of new treatments for cancer, COVID-19, and thousands of other health disorders.
“It has been a busy year at the Institute for Protein Design, designing COVID-19 therapeutics and vaccines and launching these into clinical trials, along with developing RoseTTAFold for high accuracy protein structure prediction. I am delighted that the scientific community is already using the RoseTTAFold server to solve outstanding biological problems,” said senior author David Baker, professor of biochemistry at the University of Washington School of Medicine, a Howard Hughes Medical Institute investigator, and director of the Institute for Protein Design.
In the new study, a team of computational biologists led by Baker developed the RoseTTAFold software tool. It uses deep learning to quickly and accurately predict protein structures based on limited information. Without the aid of such software, it can take years of laboratory work to determine the structure of just one protein.
RoseTTAFold, on the other hand, can reliably compute a protein structure in as little as ten minutes on a single gaming computer.
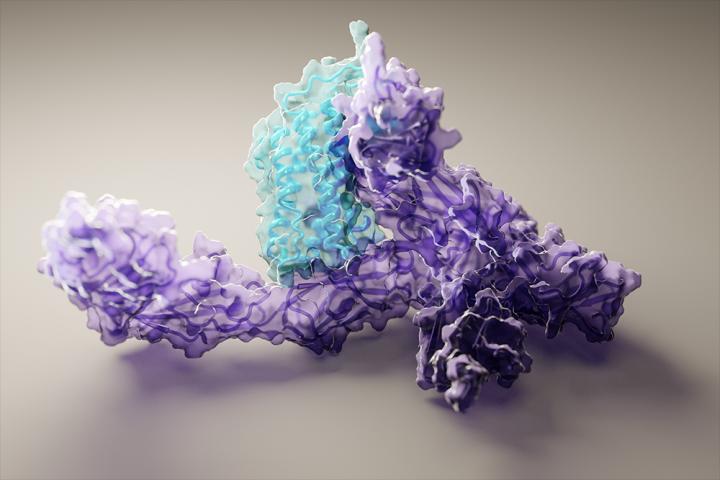
The team used RoseTTAFold to compute hundreds of new protein structures, including many poorly understood proteins from the human genome. They also generated structures directly relevant to human health, including those for proteins associated with problematic lipid metabolism, inflammation disorders, and cancer cell growth. And they show that RoseTTAFold can be used to build models of complex biological assemblies in a fraction of the time previously required.
RoseTTAFold is a “three-track” neural network, meaning it simultaneously considers patterns in protein sequences, how a protein’s amino acids interact with one another, and a protein’s possible three-dimensional structure. In this architecture, one-, two-, and three-dimensional information flows back and forth, thereby allowing the network to collectively reason about the relationship between a protein’s chemical parts and its folded structure.
“We hope this new tool will continue to benefit the entire research community,” said Minkyung Baek, a postdoctoral scholar who led the project in the Baker laboratory at UW Medicine.
###
This work was supported in part by Microsoft, Open Philanthropy Project, Schmidt Futures, Washington Research Foundation, National Science Foundation, Wellcome Trust, and the National Institute of Health. A full list of supporters is available in the Science paper.
This news release was written by Ian Haydon from the UW Medicine Institute for Protein Design.