The Scanpy software processes huge amounts of single-cell data
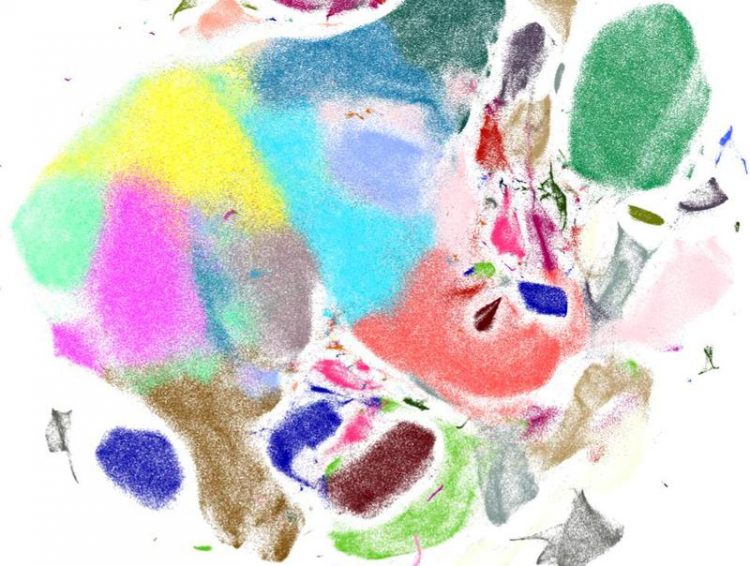
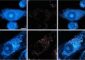
Visualization of gene expression patterns of murine brain cells generated with Scanpy Source: Helmholtz Zentrum München
“It’s about analyzing gene-expression data* of a large number of individual cells,” explains lead author Alex Wolf of the Institute of Computational Biology (ICB) at Helmholtz Zentrum München. He developed Scanpy together with his colleague Philipp Angerer in the Machine Learning Group of Prof. Dr. Dr. Fabian Theis. In addition to his position at Helmholtz Zentrum, Theis is also a professor of mathematical modelling of biological systems at the Technical University of Munich.
“New technical advances generate several orders of magnitude more data with a correspondingly greater information content,” Theis says. “However, the historically evolved software infrastructure for gene-expression analysis simply wasn’t designed to cope with the new challenges. New analytic methods are therefore needed.”
The race for the Human Cell Atlas
According to Theis, a major international research project could also benefit from the software. A team of international scientists is compiling a reference database, called the Human Cell Atlas, which holds data on the gene activity of all human cell types. “For this project, and in a growing number of other projects in which databases are combined, it is important to have scalable software,” says Theis. It is therefore no surprise that Scanpy is currently a candidate for helping to analyze the Human Cell Atlas (https://www.humancellatlas.org/).
“The publication of Scanpy marks the first software that allows comprehensive analysis of large gene-expression datasets with a broad range of machine-learning and statistical methods,” explains Wolf, describing the achievement. “The software is already being used by a number of groups around the world, notably at the Broad Institute of Harvard University and the Massachusetts Institute of Technology, MIT.”
Technologically, the application is a trailblazing development: Whereas biostatistics programs are traditionally written in the programming language R, Scanpy is based on the Python language, the dominant language in the machine learning community. Another new feature is that graph-based algorithms lie at the heart of Scanpy.
Unlike the usual approach of regarding cells as points in a coordinate system within gene-expression space, the algorithms use a graph-like coordinate system. Instead of characterizing a single cell by the expression value for thousands of genes, the system simply characterizes cells by identifying their closest neighbors – very much like the connections in social networks. In fact, to identify cell types, Scanpy uses the same algorithms as Facebook does for identifying communities.
Further information
* Expression describes how often a gene is read, i.e. it provides information on the gene’s activity.
Background:
Alex Wolf and his team have only recently occupied one of the top places in the Data Science Bowl, one of the world’s highest endowed competitions in the field of big data. For its entry, the team programmed an algorithm that can detect lung cancer within a few milliseconds on the basis of 300 layers of a three-dimensional CAT scan – a process that can take a radiologist, in the worst case, up to several hours.
In addition, the team has recently published an article in ‘Nature Communications’ on the reconstruction of cellular development processes from individual images: Paint by numbers: Algorithm reconstructs processes from individual images. https://www.helmholtz-muenchen.de/presse-medien/pressemitteilungen/alle-pressemi…
Original-Publikation:
Wolf, A. et al. (2018): Scanpy: large-scale single-cell gene expression data analysis. Genome Biology, DOI: 10.1186/s13059-017-1382-0
The Helmholtz Zentrum München, the German Research Center for Environmental Health, pursues the goal of developing personalized medical approaches for the prevention and therapy of major common diseases such as diabetes and lung diseases. To achieve this, it investigates the interaction of genetics, environmental factors and lifestyle. The Helmholtz Zentrum München is headquartered in Neuherberg in the north of Munich and has about 2,300 staff members. It is a member of the Helmholtz Association, a community of 18 scientific-technical and medical-biological research centers with a total of about 37,000 staff members. http://www.helmholtz-muenchen.de/en
The Institute of Computational Biology (ICB) develops and applies methods for the model-based description of biological systems, using a data-driven approach by integrating information on multiple scales ranging from single-cell time series to large-scale omics. Given the fast technological advances in molecular biology, the aim is to provide and collaboratively apply innovative tools with experimental groups in order to jointly advance the understanding and treatment of common human diseases. http://www.helmholtz-muenchen.de/icb
Contact for the media:
Department of Communication, Helmholtz Zentrum München – German Research Center for Environmental Health, Ingolstädter Landstr. 1, 85764 Neuherberg – Tel. +49 89 3187 2238 – Fax: +49 89 3187 3324 – E-mail: presse@helmholtz-muenchen.de
Scientific Contact:
Dr. Dr. Alexander Wolf, Helmholtz Zentrum München – German Research Center for Environmental Health, Institute of Computational Biology, Ingolstädter Landstr. 1, 85764 Neuherberg – Tel. +49 89 3187 4217, E-mail: alex.wolf@helmholtz-muenchen.de
Media Contact
All latest news from the category: Medical Engineering
The development of medical equipment, products and technical procedures is characterized by high research and development costs in a variety of fields related to the study of human medicine.
innovations-report provides informative and stimulating reports and articles on topics ranging from imaging processes, cell and tissue techniques, optical techniques, implants, orthopedic aids, clinical and medical office equipment, dialysis systems and x-ray/radiation monitoring devices to endoscopy, ultrasound, surgical techniques, and dental materials.
Newest articles

A universal framework for spatial biology
SpatialData is a freely accessible tool to unify and integrate data from different omics technologies accounting for spatial information, which can provide holistic insights into health and disease. Biological processes…

How complex biological processes arise
A $20 million grant from the U.S. National Science Foundation (NSF) will support the establishment and operation of the National Synthesis Center for Emergence in the Molecular and Cellular Sciences (NCEMS) at…

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…