Patents for Protein Biomarker Discovery

Peakadilly receives multiple patents for breakthrough Protein biomarker discovery and identification technologies.
The patent offices of the United States of America and Canada have granted patents for Peakadilly’s core technology, Cofradic. Cofradic, initially developed at the University of Ghent, Belgium, allows the identification and quantification of proteins in any biological sample and represents the most important component of the company’s Masstermind biomarker discovery engine. Peakadilly has already received patents for the technology in Europe.
Peakadilly’s proprietary proteomics technologies allow fast, automated and highly flexible and sensitive qualitative and quantitative proteomics without using gels and without the use of an affinity tag. The technology comprises a suite of chemistries and chromatographic methods to reduce the natural complexity of any protein mixture. Together with a novel serum depletion technology and state-of-the art mass spectrometry, Peakadilly’s Masstermind platform allows to analyse huge sets of proteins and posttranslational modified proteins in clinical samples like blood or sputum, offering unrivalled sensitivities and high content data unseen with genomics & transcriptomics. Using software tools developed in-house, the quantitative comparison of hundreds of samples is possible, making the technology suited for biomarker discovery within clinical trials. In collaboration with a big pharma company, the technology is currently being applied to distinguish responders from non-responders in a specific therapeutic regimen.
Peakadilly is a Belgian biotech company focused on the discovery and development of next generation molecular diagnostics that will revolutionize the development of innovative treatments for cancer and inflammatory diseases, and that will make health care more personalized and predictive. The company was founded late 2004 by Dr. Koen Kas, Dr. Joel Vandekerckhove and VIB as a start-up from the Flanders Interuniversity Institute for Biotechnology (VIB) and Ghent University and today employs 12 people. Apart from research collaborations, Peakadilly provides access to its technology also in fee-for-service contracts.
Media Contact
More Information:
http://www.peakadilly.comAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
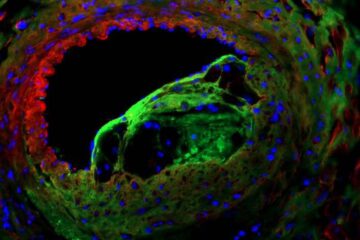
Solving the riddle of the sphingolipids in coronary artery disease
Weill Cornell Medicine investigators have uncovered a way to unleash in blood vessels the protective effects of a type of fat-related molecule known as a sphingolipid, suggesting a promising new…

Rocks with the oldest evidence yet of Earth’s magnetic field
The 3.7 billion-year-old rocks may extend the magnetic field’s age by 200 million years. Geologists at MIT and Oxford University have uncovered ancient rocks in Greenland that bear the oldest…
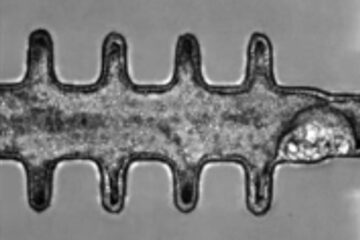
Mini-colons revolutionize colorectal cancer research
As our battle against cancer rages on, the quest for more sophisticated and realistic models to study tumor development has never been more critical. Until now, research has relied on…





















