Newly discovered deep-sea microbes gobble greenhouse gases and perhaps oil spills, too
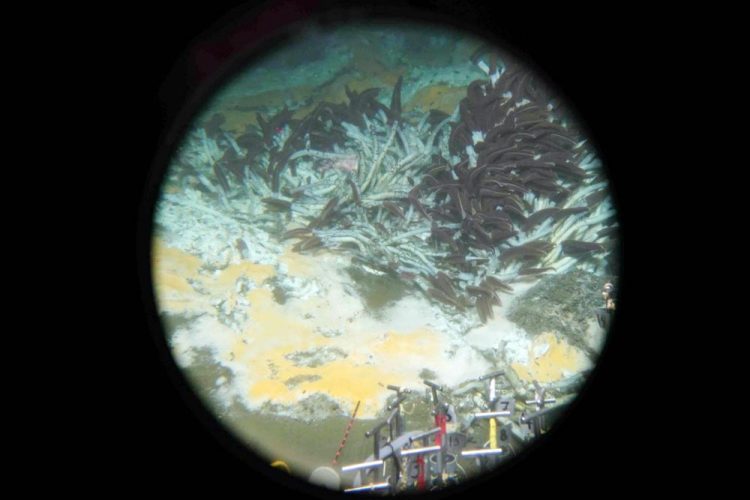
Researchers have documented extensive diversity in the microbial communities living in the extremely hot, deep-sea sediments located in the Guaymas Basin in the Gulf of California. This view of the Guaymas Basin seafloor was taken through the window of the Alvin submersible by Brett Baker in November 2018. Credit: Brett Baker/University of Texas at Austin
In a paper published in Nature Communications this week, researchers documented extensive diversity in the microbial communities living in the extremely hot, deep-sea sediments located in the Guaymas Basin in the Gulf of California.
The team uncovered new microbial species that are so genetically different from those that have been previously studied that they represent new branches in the tree of life. Many of these same species possess keen pollutant-eating powers, like other, previously identified microbes in the ocean and soil.
“This shows the deep oceans contain expansive unexplored biodiversity, and microscopic organisms there are capable of degrading oil and other harmful chemicals,” said assistant professor of marine science Brett Baker, the paper's primary investigator.
“Beneath the ocean floor huge reservoirs of hydrocarbon gases–including methane, propane, butane and others–exist now, and these microbes prevent greenhouse gases from being released into the atmosphere.”
The new study, representing the largest-ever genomic sampling of Guaymas Basin sediments, was co-authored by former UT postdoctoral researcher Nina Dombrowski and University of North Carolina professor Andreas P. Teske.
The researchers' analysis of sediment from 2,000 meters below the surface, where volcanic activity raises temperatures to around 200 degrees Celsius, recovered 551 genomes, 22 of which represented new entries in the tree of life.
According to Baker, these new species were genetically different enough to represent new branches in the tree of life, and some were different enough to represent entirely new phyla.
“The tree of life is something that people have been trying to understand since Darwin came up with the concept over 150 years ago, and it's still this moving target at the moment,” said Baker, who earlier was part of a team that mapped the most comprehensive genomic tree of life to date.
“Trying to map the tree is really kind of crucial to understanding all aspects of biology. With DNA sequencing and the computer approaches that we use, we're getting closer, and things are expanding quickly.”
Only about 0.1 percent of the world's microbes can be cultured, which means there are thousands, maybe even millions, of microbes yet to be discovered.
Baker's team investigates interactions between microbial communities and the nutrients available to them in the environment by taking samples of sediment and microbes in nature, and then extracting DNA from the samples. The researchers sequence the DNA to piece together individual genomes, the sets of genes in each organism, and infer from the data how microbes consume different nutrients.
“For this, we try to look for organisms that have been studied before and look for similarities and differences,” said Dombrowski, who is now at the Royal Netherlands Institute for Sea Research. “This might initially sound easy, but really is not, since often more than half of the genes we find are so far uncharacterized and unknown.”
The samples were collected using the Alvin submersible, the same sub that found the Titanic, because the microbes live in extreme environments. Teske, who collaborated with Baker and Dombrowski, has driven sample collection at Guaymas Basin for several years, working with scientists across the world who are using different approaches to study life there.
This month, Baker is part of a team on the Alvin sampling in areas of the basin that previously have never been studied.
“We think that this is probably just the tip of the iceberg in terms of diversity in the Guaymas Basin,” Baker said. “So, we're doing a lot more DNA sequencing to try to get a handle on how much more there is. This paper is really just our first hint at what these things are and what they are doing.”
###
This research was supported by the U.S. Department of Energy, the Sloan Foundation and the U.S. National Science Foundation.
Once the paper is published, it will be available at: http://dx.
Media Contact
More Information:
http://dx.doi.org/10.1038/s41467-018-07418-0All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Sea slugs inspire highly stretchable biomedical sensor
USC Viterbi School of Engineering researcher Hangbo Zhao presents findings on highly stretchable and customizable microneedles for application in fields including neuroscience, tissue engineering, and wearable bioelectronics. The revolution in…

Twisting and binding matter waves with photons in a cavity
Precisely measuring the energy states of individual atoms has been a historical challenge for physicists due to atomic recoil. When an atom interacts with a photon, the atom “recoils” in…
Nanotubes, nanoparticles, and antibodies detect tiny amounts of fentanyl
New sensor is six orders of magnitude more sensitive than the next best thing. A research team at Pitt led by Alexander Star, a chemistry professor in the Kenneth P. Dietrich…





















