Ten thousand bursting genes
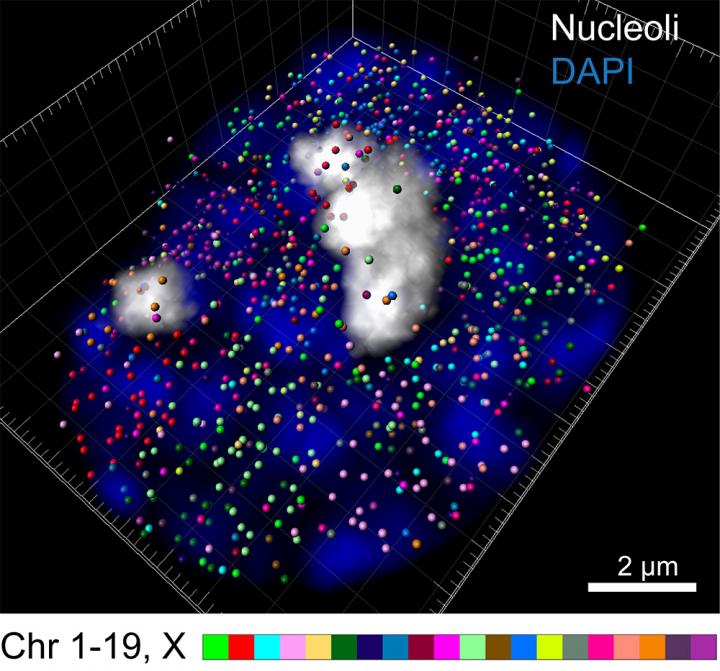
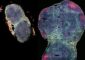
Intron seqFISH enables 3D reconstruction of nascent transcription active sites (colored spots) in an embryonic stem cell (blue), with individual chromosomes occupying distinct spatial territories (colored differently). Here, 982 transcription active sites, corresponding to individual genes, are present in this cell. Credit: Cai laboratory / Cell
A breakthrough new technique enables scientists to image 10,421 genes at once within individual cells.
The work was done in the laboratory of Long Cai, research professor in biology and an affiliated faculty member of the Tianqiao and Chrissy Chen Institute for Neuroscience at Caltech. A paper describing the research appears in the June 7 issue of the journal Cell.
The new technique, dubbed intron seqFISH (sequential fluorescence in situ hybridization), is a major advance in being able to identify what goes on across the genome in hundreds of different cells at once. Previously, researchers could only image four to five genes at a time in cells with microscopy. This work builds off of previous advances from the Cai laboratory, including an earlier version of seqFISH from 2014 and research from 2017 that profiled over 10,000 genes under a microscope. Scaling seqFISH up to a genomic level now enables the imaging of over 10,000 genes–about half of the total number of genes in mammals–within single cells.
In order for genetic instructions to be turned into an actual functioning protein, a process called transcription must first occur. This process often occurs in pulses, or “bursts.” First, a gene will be read and copied into a precursor messenger RNA, or pre-mRNA, like jotting a quick, rough draft. This molecule then matures into a messenger RNA, or mRNA, akin to editing the rough draft. During the “editing” process, certain regions called introns are cut out of the pre-mRNA.
The team chose to focus on labeling introns because they are produced so early in the transcription process, giving a picture of what a cell is doing at the precise moment of gene expression.
Using the newly developed intron seqFISH technique, each intron is labeled with a unique fluorescent barcode, enabling it to be seen with a microscope. Seeing introns reveals which genes are currently turned on in individual cells, how strongly they are expressed, and where they are located. 10,421 introns–and therefore 10,421 genes–can be imaged at once.
Previous work that developed the barcoding technique focused on labeling mRNA itself, providing a measurement of how gene expression changed over several hours as the mRNA developed. Looking at introns enabled the researchers to examine, for the first time, so-called nascent transcriptomes–newly synthesized gene expression. This led them to discover that the transcription of genes oscillates globally across many genes on what Cai calls a “surprisingly short” timescale–only about two hours–compared to the time it takes for a cell to divide and replicate itself, which takes from 12 to 24 hours. This means that over the course of a two-hour period, many genes within a cell will burst on and off.
There are several reasons why the oscillation phenomenon had not been observed previously. First, because these two-hour oscillations are not synchronized amongst different cells, the fluctuations are averaged out by methods that require many cells. Second, the high accuracy of the seqFISH method allows the researchers to be certain that what they observe represents real biological fluctuations, rather than technical noise. Lastly, these two-hour oscillations are obscured when mRNAs rather than introns are measured, because mRNA molecules have a longer lifetime, three to four hours, in mammalian cells.
Additionally, because introns stay where the gene is physically located, fluorescently imaging introns allows researchers to visualize where genes are located within the chromosome, the large structure that DNA folds into within the cell's nucleus. In this work, the team was surprised to discover that most active, protein-encoding genes are located on the surface of the chromosome, not buried inside of it.
“This technique can be applied to any tissue,” says Cai, who is a collaborator on the Human Cell Atlas, a project that aims to define all cell types in the human body. “Intron seqFISH can help identify cell types and also what the cells are going to do, in addition to giving us a look at the chromosome structure in the same cells.”
###
The paper is titled “Dynamics and spatial genomics of the nascent transcriptome in single cells by intron seqFISH.” Former graduate student Sheel Shah (PhD '17) and current graduate students Wen Zhou and Yodai Takei are the study's first authors. Additional co-authors include graduate student Chee-Huat Linus Eng, former graduate student Eric Lubeck (PhD '16), research technicians Jina Yun and Noushin Koulena, graduate student Eric Liaw, visiting student Mina Amin, as well as fluidics and mechanical engineers Christopher Cronin and Chris Karp. Funding was provided by the Nakajima Foundation, the National Institutes of Health, and the Paul G. Allen Foundation Discovery Center.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Red light therapy for repairing spinal cord injury passes milestone
Patients with spinal cord injury (SCI) could benefit from a future treatment to repair nerve connections using red and near-infrared light. The method, invented by scientists at the University of…
Insect research is revolutionized by technology
New technologies can revolutionise insect research and environmental monitoring. By using DNA, images, sounds and flight patterns analysed by AI, it’s possible to gain new insights into the world of…

X-ray satellite XMM-newton sees ‘space clover’ in a new light
Astronomers have discovered enormous circular radio features of unknown origin around some galaxies. Now, new observations of one dubbed the Cloverleaf suggest it was created by clashing groups of galaxies….