Protein Sequences: Not So Predictable After All

Scientists have believed for decades that the sequencing of the human genome would automatically yield the sequences of proteins, the functional products of genes, and thus lead to the unraveling of the mechanisms behind human cell biology and disease. However, a paper published in Science today by the Ludwig Institute for Cancer Research (LICR) describes a novel cellular process that casts some doubt on the accepted paradigm of deducing a protein’s sequence from the DNA sequence of its gene.
When a protein is called upon to act in a cellular process, segments (exons) of the gene’s DNA are transcribed into RNA fragments, which are then spliced (joined) together according to directions encoded by the DNA. The RNA transcript is then translated into amino acids which form a protein. When the protein is no longer required by the cell, it is degraded by a specialized complex called the ‘proteasome’. Although functionally different proteins can be produced from one gene sequence – by modification through the addition of small molecules to the amino acids, by truncation of the sequence, or by the translation of RNA transcripts that omit some exons – they are variations on a theme determined essentially by the gene’s DNA sequence. However scientists at the LICR Branch in Brussels, Belgium, have now shown that the sequence of a human protein can actually be altered by the proteasome in ways that are completely unpredicted by the gene sequence. The discovery is both a new process for protein processing and a novel, hitherto unsuspected role for the proteasome.
‘Post-translational splicing’ was discovered when the LICR team analyzed small protein sequences that activate the immune system in response to the presence of cancer cells. These ‘peptide antigens’ are created when the proteasome fragments a cancer-specific protein into peptides, which are then transported to the cell surface to mark the cell for immunological destruction. The team found that one peptide antigen, which stimulated the immune system to recognize and destroy melanoma cells, had a sequence quite unlike that predicted by its gene sequence. Upon investigating further, the researchers found that the proteasome had cut a peptide in three and had then rejoined the pieces so that the peptide antigen now recognized by the immune system was missing part of its original sequence.
“Post-translational splicing is a fascinating process in its own right,’ says Dr. Benoît Van den Eynde, the senior author of the study, “but it also has important implications for the design of cancer or HIV vaccines based on peptide antigens. Synthetic peptides being investigated in early-phase clinical trials of vaccines are usually designed based on the gene sequence of cancer-specific proteins. However post-translational splicing may be modifying the peptide antigens in ways that we haven’t even suspected before now.”
The issue of predicted versus post-translationally spliced, novel peptide antigens is a particularly important consideration when monitoring immunological responses to cancer vaccines in patients. The existing methodologies typically quantify the presence of T cells specific for the predicted peptide antigen, and would not detect T cells specific for the novel peptide antigen. Additional studies are required to determine if proteome-mediated post-translational splicing occurs in relation to cancer and/or virally-infected cells, and if so, the frequency of occurrence of such novel peptide antigens in these diseases.
The discovery of post-translational splicing has immunologists intrigued by the additional complications in antigen identification, and cell biologists excited by a new insight into the proteasome’s bag of tricks. However for geneticists and biochemists, the discovery of post-translational splicing may have produced a slight sense of unease. Until we know exactly when, why, and how often post-translational splicing occurs, we can no longer automatically assume that the human genome holds all the answers for predicting protein sequences.
Media Contact
More Information:
http://www.licr.orgAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Why getting in touch with our ‘gerbil brain’ could help machines listen better
Macquarie University researchers have debunked a 75-year-old theory about how humans determine where sounds are coming from, and it could unlock the secret to creating a next generation of more…

Attosecond core-level spectroscopy reveals real-time molecular dynamics
Chemical reactions are complex mechanisms. Many different dynamical processes are involved, affecting both the electrons and the nucleus of the present atoms. Very often the strongly coupled electron and nuclear…
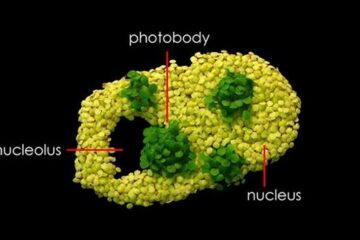
Free-forming organelles help plants adapt to climate change
Scientists uncover how plants “see” shades of light, temperature. Plants’ ability to sense light and temperature, and their ability to adapt to climate change, hinges on free-forming structures in their…