Mapping the 'dark matter' of human DNA
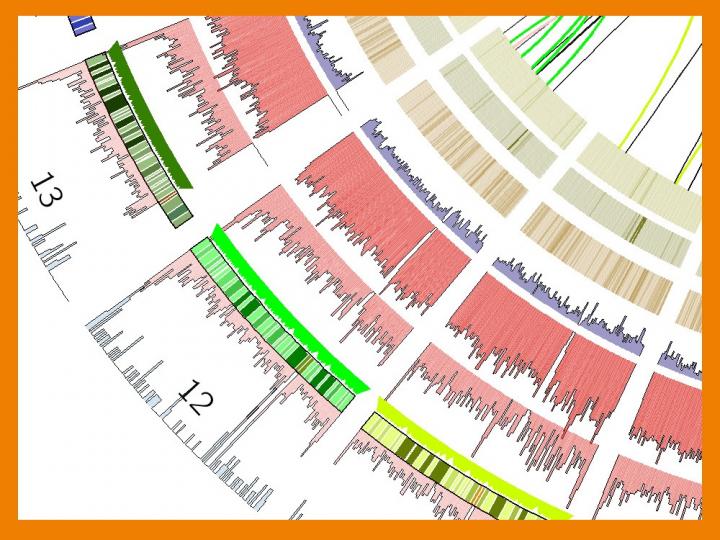
This is a detail of a representation of a DNA variants map. Credit: Nature Communications (DOI: 10.1038/ncomms12989)
Although our knowledge of the human DNA is extensive, it is nowhere near complete. For instance, our knowledge of exactly which changes in our DNA are responsible for a certain disease is often insufficient. This is related to the fact that no two people have exactly the same DNA. Even the DNA molecules of identical twins have differences, which occur during their development and ageing.
Some differences ensure that not everybody looks exactly alike, while others determine our susceptibility to particular diseases. Knowledge about the DNA variants can therefore tell us a lot about potential health risks and is a first step towards personalized medicine.
Many small variants in the human genome – the whole of genetic information in the cell – have already been documented. Although it is known that larger structural variants play an important role in many hereditary diseases, these variants are also more difficult to detect and are, therefore, much less investigated.
By comparing the DNA of 250 healthy Dutch families with the reference DNA database the researchers were able to identify 1.9 million variants affecting multiple DNA 'letters'. These variants include large sections of DNA that have disappeared, moved or even appear out of nowhere.
When this happens in the middle of a gene that encodes a certain protein, it is likely that the functionality of the gene, and thus the production of the protein, is compromised. However, large structural variants often occur just before or after the coding part of a gene. The effect of this type of variation is hard to predict.
In the paper two occasions are described in which an extra piece of DNA was found just outside the coding region of a gene. In these occasions the variants had a demonstrable effect on the gene regulation. This proves that even structural variants that occur outside the coding regions need to be monitored closely in future DNA screenings.
The catalogue of variants provided by this research enables other scientists to predict the occurrence of large structural variants from the known profile of the smaller ones. This technique opens new possibilities for studying the effects of large structural changes in our genomes.
Additionally, the research resulted in the discovery of large parts of DNA that were not included in the genome reference. This “extra” DNA does contain parts that could be involved in the production of proteins. One of the extra pieces of DNA that was described in the paper is a new “ZNF” gene that has previously never been found in humans. Nevertheless it appears to be present in roughly half of the Dutch population.
This particular gene is a member of the ZNF gene family that was known from the reference genomes of several species of apes. The new variant will now be added to the human reference database. Authors subsequently showed that this gene is also present in genomes of several other human populations, however its function remains unknown. The fact that these and other pieces of “dark matter” now have been placed on the genetic map enables scientists worldwide to study them and use the results to better understand human genetic diseases.
This study is part of the Genome of the Netherlands (GoNL) project. One of the main goals of the study is to map the genome of the Dutch population and all its variants. Several teams of bio- informaticians from different countries work continuously on the development of new algorithms for data analysis, as well as on innovative ways to combine existing algorithms. The result: an accurate representation of the genomes of the Dutch population and thereby a solid base for the personalised medicine of the future.
###
“A high-quality human reference panel reveals the complexity and distribution of genomic structural variants”: https:/
For more information on GoNL: http://www.
ERIBA: European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands
Radboud UMC: Department of Human Genetics, Donders Institute, Radboud University Medical Center, Nijmegen, The Netherlands
XJTU: Xi'an Jiaotong University, Xi'an, China
Saarland University: Center for Bioinformatics, Saarland University, Saarbrücken, Germany
CWI: Life Sciences Group, Centrum Wiskunde & Informatica, Amsterdam, The Netherlands.
UMC Utrecht: Center for Molecular Medicine, Division of Biomedical Genetics, University Medical Center Utrecht, Utrecht, The Netherlands
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Sea slugs inspire highly stretchable biomedical sensor
USC Viterbi School of Engineering researcher Hangbo Zhao presents findings on highly stretchable and customizable microneedles for application in fields including neuroscience, tissue engineering, and wearable bioelectronics. The revolution in…

Twisting and binding matter waves with photons in a cavity
Precisely measuring the energy states of individual atoms has been a historical challenge for physicists due to atomic recoil. When an atom interacts with a photon, the atom “recoils” in…
Nanotubes, nanoparticles, and antibodies detect tiny amounts of fentanyl
New sensor is six orders of magnitude more sensitive than the next best thing. A research team at Pitt led by Alexander Star, a chemistry professor in the Kenneth P. Dietrich…





















