Largest, most accurate list of RNA editing sites
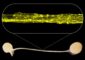
The two two dark humps in the top row (representing fly RNA) are missing in flies that lack the enzyme to edit RNA (middle row) and missing in the DNA of normal flies. The analysis suggests they are sites of RNA editing.<br><br>Credit: Reenan lab/Brown University<br>
The “master list” totals 3,581 sites in which the enzyme ADAR might swap an “A” nucleotide for a “G” in an RNA molecule. Such a seemingly small tweak means a lot because it changes how genetic instructions in DNA are put into action in the fly body, affecting many fundamental functions including proper neural and gender development. In humans, perturbed RNA editing has been strongly implicated in the diseases ALS and Acardi-Gutieres disease.
The new list of editing sites could therefore help thousands of researchers studying the RNA molecules that are transcribed from DNA, the so-called “transcriptome,” by providing reliable information about the thousands of editing changes that can occur.
“Drosophila serves as a model for all the organisms where people are studying transcriptomes,” said the paper's corresponding author Robert Reenan, professor of biology in the Department of Molecular Biology, Cell Biology, and Biochemistry at Brown. “But in the early days of RNA editing research, the catalog of these sites was determined completely by chance – people working on genes of interest would discover a site. The number of sites grew slowly.”
In fact, Reenan was co-author of a paper in Science 10 years ago that made a splash with only 56 new editing sites which at the time, more than doubled the number of known sites in the entire field.
Validation means accuracy
Several more recent attempts to catalog RNA editing sites have yielded larger catalogs, but those contained many errors (the paper provides a comparison between the new list and previous efforts such as ModENCODE).
To avoid such mistakes, Reenan and colleagues, including lead author and graduate student Georges St. Laurent, painstakingly validated 1,799 of the sites. They worked with Charles Lawrence, professor of applied mathematics and the paper's co-senior author, to predict another 1,782 sites and validated a statistically rigorous sampling of those.
In all, the team's methodology allowed them to estimate that the combined list of 3,581 directly observed and predicted sites is 87 percent accurate.
“The sites that we validated, for anyone who wants to do the same experiment under the same conditions, the sites should be there,” said co-author and postdoctoral researcher Yiannis Savva. “In other papers, they just did sequencing to say there is an editing site there, but when you check, it's not there.”
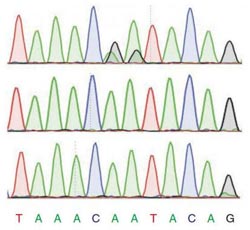
The researchers used the tried-and-true, decades-old Sanger method of sequencing to double-check all the candidate editing sites that they had found using the high-throughput technology called single molecule sequencing. They compared the sequenced RNA of a population of fruit flies to their sequenced DNA and to the RNA of another population of flies engineered to lack the ADAR editing enzyme. By comparing these three sequences they were able to see the A-to-G changes that could not be attributed to anomalies in DNA (i.e., mutations, or single-nucleotide polymorphisms) and that never occurred in flies incapable of editing.
As they conducted their validations, they fed the results back into their prediction algorithm. Over several iterations, that computer model “learned” to make better and better predictions. They ultimately found 77 different variables that helped them to distinguish real editing sites from nucleotides that were conclusively not editing sites.
Biological insights
The researchers then examined the implications of the patterns they saw in their data and gained several insights.
One was that a considerable amount of editing occurs in sections of RNA that do not code for making proteins. Editing is concentrated in a small number of RNAs, raising the question, Lawrence said, of what accounts for that selectivity.
“How does the cell go about choosing which ones are going to get edited and which aren't is an interesting question this opens,” he said.
Where editing is found, the researchers discovered, there is usually more alternative splicing, which means the body is more often assembling a different recipe from its genetic instructions to make certain proteins.
The researchers also found that the RNAs that are most heavily edited tend to be expressed to a lesser extent, decreasing how often they are put into action in the body.
RNA editing helps explain why organisms are even more different from each other – and from themselves at different times — than DNA differences alone would suggest.
“RNA editing has emerged as a way to diversify not just the proteome but the transcriptome overall,” Reenan said.
In addition to Reenan, Lawrence, Savva, and St. Laurent, who is also affiliated with the St. Laurent Institute in Cambridge, Mass., the paper's other authors are Michael Tackett, Sergey Nechkin, Dimitry Shtokalo, and Philipp Kapranov of the St. Lawernce Institute, Denis Antonets of the State Research Center of Virology and Biotechnology in Russia, and Rachael Maloney, a Brown graduate now at the University of Massachusetts Medical School.
Reenan received funding from the Ellison Medical Research Foundation.
Media Contact
More Information:
http://www.brown.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…