Newly discovered group of algae live in both fresh water and ocean
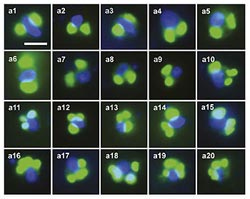
A collection of rappemonad cells photographed by a high-powered microscope. Each cell contains at least two chloroplasts (green dots) and a nucleus (blue dots). Images: from Kim, Harrison, Sudek et al. PNAS 2010 <br>
A team of biologists has discovered an entirely new group of algae living in a wide variety of marine and freshwater environments. This group of algae, which the researchers dubbed “rappemonads,” have DNA that is distinctly different from that of other known algae.
In fact, humans and mushrooms are more closely related to each other than rappemonads are to some other common algae (such as green algae). Based on their DNA analysis, the researchers believe that they have discovered not just a new species or genus, but a potentially large and novel group of microorganisms.
The rappemonads were found in a wide range of habitats, in both fresh and salt water, and at temperatures ranging from 52 degrees to 79 degrees Fahrenheit. According to MBARI Senior Research Technician Sebastian Sudek, co-first-author of the paper reporting the discovery of these algae, “Based on the evidence so far, I think it's fair to say that rappemonads are likely to be found throughout many of the world's oceans. We don't know how common they are in fresh water, but our samples were not from unusual sources—they were from small lakes and reservoirs.”
Researchers Sebastian Sudek, Heather Wilcox, and Alexandra Worden of the Monterey Bay Aquarium Research Institute (MBARI), along with collaborators at Dalhousie University and the Natural History Museum (NHM), London, discovered these microscopic algae by following up on an unexpected DNA sequence listed in a research paper from the late 1990s. They named the newly identified group of algae “rappemonads” after Michael Rappé, a professor at the University of Hawaii who was first author of that paper.
Following up on their initial lead, the research team developed two different DNA “probes” that were designed to detect the unusual DNA sequences reported by Rappé. Using these new probes, the researchers analyzed samples collected by Worden’s group from the Northeast Pacific Ocean, the North Atlantic, the Sargasso Sea, and the Florida Straits, as well as samples collected from several freshwater sites by co-author Thomas Richards' group at NHM. To the teams’ surprise, they discovered evidence of microscopic organisms containing the unusual DNA sequence at all five locations.
Although the rappemonads were fairly sparse in many of the samples, they appear to become quite abundant under certain conditions. For example, water samples taken from the Sargasso Sea near Bermuda in late winter appeared to have relatively high concentrations of rappemonads.
When asked why these apparently widespread algae had not been detected sooner, Sudek speculates that it may in part be due to their size. “They are too small to be noticed by people who study bigger algae such as diatoms, yet they may be filtered out by researchers who study the really small algae, known as picoplankton.”
Sudek says, “The rappemonads are just one of many microbes that we know nothing about—this makes it an exciting field in which to work.” Worden, in whose lab the research was conducted, and who first noticed the unique sequence in the 1990 paper, then initiated research to “chase down” the story behind that sequence, continues, “Right now we treat all algae as being very similar. It is as if we combined everything from mice up to humans and considered them all to have the same behaviors and influence on ecosystems. Clearly mice and humans have different behaviors and different impacts!”
Even though DNA analysis demonstrated that rappemonads were present in their water samples, the researchers were still unable to visualize the tiny organisms because they didn't know what physical characteristics to look for. However, by attaching fluorescent compounds to the newly developed DNA probes, and then applying these probes to intact algae cells, Eunsoo Kim at Dalhousie was able to make parts of the rappemonads glow with a greenish light. This allowed the researchers to see individual rappemonads under a microscope.
The greenish glow highlighted the rappemonad’s “chloroplasts,” which contain the unique DNA sequence tagged by the new probes. Chloroplasts are used by plants and algae to harvest energy from sunlight in a process called photosynthesis. Because all of the rappemonads contain chloroplasts, the researchers believe they “make a living” through photosynthesis. However, Worden points out that it still needs to be shown that the chloroplasts are functional.
One of the primary goals of Worden’s research is to study marine algae in the context of their environment. Worden feels that such an approach is imperative to understanding how rappemonads and other microorganisms affect large-scale processes in the ocean and in the atmosphere. In coming years her lab will be building upon their recent insights, including the discovery of the rappemonads, to study the roles that different algal groups play in the cycling of carbon dioxide between the atmosphere and the ocean.
Worden says, “There is a tremendous urgency in gaining an understanding of biogeochemical cycles. Marine algae are key players in these cycles, taking up carbon dioxide from the atmosphere and releasing oxygen, which we breathe. Until we have a true census of marine algae and understanding of how each group thrives, it will be very difficult to model global biogeochemical cycles. Such modeling is essential for predicting how climate change will impact life on earth.”
For additional information or images relating to this news release, please contact:
Kim Fulton-Bennett: (831) 775-1835, kfb@mbari.org
Research paper:
E. Kim, J.W. Harrison, S. Sudek, M.D.M. Jones, H.M. Wilcox, T.A. Richards, A.Z. Worden, J.M. Archibald, Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proceedings of the National Academy of Sciences (PNAS), DOI: 10.1073/pnas.1013337108.
Media Contact
All latest news from the category: Ecology, The Environment and Conservation
This complex theme deals primarily with interactions between organisms and the environmental factors that impact them, but to a greater extent between individual inanimate environmental factors.
innovations-report offers informative reports and articles on topics such as climate protection, landscape conservation, ecological systems, wildlife and nature parks and ecosystem efficiency and balance.
Newest articles

A universal framework for spatial biology
SpatialData is a freely accessible tool to unify and integrate data from different omics technologies accounting for spatial information, which can provide holistic insights into health and disease. Biological processes…

How complex biological processes arise
A $20 million grant from the U.S. National Science Foundation (NSF) will support the establishment and operation of the National Synthesis Center for Emergence in the Molecular and Cellular Sciences (NCEMS) at…

Airborne single-photon lidar system achieves high-resolution 3D imaging
Compact, low-power system opens doors for photon-efficient drone and satellite-based environmental monitoring and mapping. Researchers have developed a compact and lightweight single-photon airborne lidar system that can acquire high-resolution 3D…





















