The MiSIng Piece Revealed: Classifying Microbial Species in the Genomics Era
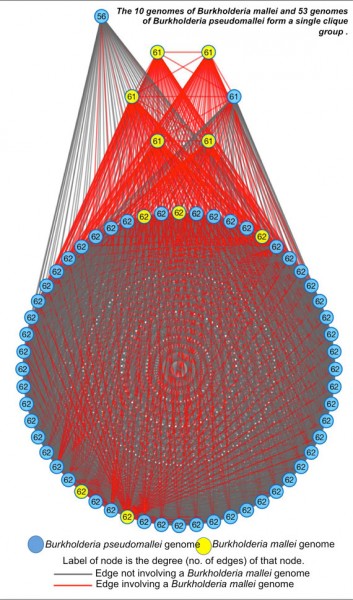
Figure by Neha Varghese The clique-group comprising of pathogenic Burkholderia mallei and Burkholderia pseudomallei strains highlights the ability of MiSI to capture high similarity as well as subtle differences between closely related genomes.
Traditionally, identifying the microorganisms selected for sequencing is often decided on the basis of a single universal marker gene. More recently, however, researchers have noticed that the identity of microbes for which whole-genome information has become available does not always match up with the identity determined by the approaches commonly used prior to the advent of next-generation high-throughput sequencing.
In a study published ahead online July 6, 2015 in Nucleic Acids Research (NAR), a team of researchers from the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility and their collaborators developed and evaluated a new method for classifying microbial species that could be supplemented – as needed – by traditional approaches relied on by microbiologists for decades. Study first author Neha Varghese of the DOE JGI says the Microbial Species Identifier (MiSI) method meets a “long standing need for a systematic, scalable, and objective microbial species assignment technique.”
“A fast, genome-sequence based method”
The standard whole-genome approach relies on the small subunit ribosomal RNA gene (16S rRNA); without sequencing, researchers used approaches such as DNA-DNA hybridization, phenotypic information – genotyping, phenotyping, or classifying by the chemical compounds that microbes share – to derive the needed information for microbial classification.
The MiSI method developed at the DOE JGI relies primarily on genome sequencing and is a combination of two metrics for determining how closely related two genomes are: genome-wide Average Nucleotide Identity (gANI) and alignment fraction (AF). Computational tool development is a critical element of the DOE JGI 10-Year Strategic Vision, essential to characterizing complex biological and environmental systems in support DOE’s research missions, as well as the Institute’s partnership with the National Energy Research Scientific Computing Center (NERSC).
“Scientists and practitioners of microbiology will much appreciate the robust, extensive, taxonomic organization of the microbial world provided by this fast, genome-sequence based method,” said Jim Tiedje, Director of the Center for Microbial Ecology at Michigan State University. “It provides a more accurate and clear-cut means to identify bacteria.” A DOE JGI collaborator on the Great Prairie Soil Metagenome Grand Challenge project, Tiedje and his former student Kostas Konstantinidis, one of the study’s co-authors, jointly developed the original genome-wide gANI metric modified by the DOE JGI team as a basis for the MiSI method.
The algorithm in the gANI method developed by Tiedje and Konstantinidis used segments sampled over the whole genome and the National Center for Biotechnology Information (NCBI) tool Basic Local Alignment Search Tool (BLAST) for sequence alignment. MiSI speeds up the computations dramatically—by about 10-fold—by using nucleotide sequences of genes and a modified BLAST-based similarity search.
The team implemented the MiSI method over a massive database of more than 13,000 bacterial and archaeal high quality genomes selected from the Integrated Microbial Genomes (IMG) database. These genomes were then classified into clusters represented by cliques or clique-groups where connectivity is determined by genomic similarity (a surrogate for evolutionary distance), and thus, for the first time, allowed researchers to diagram how genomes are related to each other across a large phylogenetic space.
The completely connected nature of these cliques helped the team identify highly conserved cores of species, while the semi-connect nature of clique groups helped identify species that could be revisited taxonomically. Further, since the clustering is based solely on genomic relatedness, the team was able to use this method to determine if an uncultured organism is either related to an existing species or is a novel candidate species.
“A universal method for species identification”
“The implications of using this method in transforming microbiology cannot be overstated,” said Kyrpides, head of the Prokaryote Super Program and co-corresponding author in this study. “We now have a universal method for species identification across all Archaea and Bacteria that relies on the entire genome rather than a single gene, or a small number of marker genes. When applied to all the sequenced genomes currently available, this method enabled us to observe species evolution in action, manifested through what we call clique groups. Perhaps one of the most dramatic observations we had was that more than half of all the species that had more than one sequenced genome, had at least one genome wrongly named.”
Kyrpides went on to say, “This method is also shedding light on the long debated issue of microbial species. The fact that over 86% of all the microbial species for which more than one genome has been sequenced are grouping into separate cliques, strongly supports the notion of a microbial species, as opposed to the idea of genetic continuum among microbial species, which however was observed for a small number of species, about 5%. It would be very interesting to see how these observations evolve as the numbers of microbial genomes sequenced explode.”
The DOE JGI sequencing pipeline is already using the MiSI method to determine the similarity of newly sequenced genomes to existing reference genomes. MiSI is available for use by the general research community through the IMG system and the data used by the DOE JGI team for this study are publicly available at https://ani.jgi-psf.org .
The U.S. Department of Energy Joint Genome Institute, User Facility of Lawrence Berkeley National Laboratory supported by the DOE Office of Science, is committed to advancing genomics in support of DOE missions related to clean energy generation and environmental characterization and cleanup. DOE JGI, headquartered in Walnut Creek, Calif., provides integrated high-throughput sequencing and computational analysis that enable systems-based scientific approaches to these challenges. Follow @doe_jgi on Twitter.
DOE’s Office of Science is the largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.
Contact Information
David Gilbert
Public Affairs Manager
U.S. Department of Energy Joint Genome Institute
degilbert@lbl.gov
Phone: 925-296-5643
Media Contact
More Information:
http://www.science.doe.govAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Sea slugs inspire highly stretchable biomedical sensor
USC Viterbi School of Engineering researcher Hangbo Zhao presents findings on highly stretchable and customizable microneedles for application in fields including neuroscience, tissue engineering, and wearable bioelectronics. The revolution in…

Twisting and binding matter waves with photons in a cavity
Precisely measuring the energy states of individual atoms has been a historical challenge for physicists due to atomic recoil. When an atom interacts with a photon, the atom “recoils” in…
Nanotubes, nanoparticles, and antibodies detect tiny amounts of fentanyl
New sensor is six orders of magnitude more sensitive than the next best thing. A research team at Pitt led by Alexander Star, a chemistry professor in the Kenneth P. Dietrich…





















