Massive project reveals shortcomings of modern genome analysis

“We've made huge progress in recent years with many genomes, including humans, but a lot of the problems can't be solved by simply dumping data into a computer and having truth and light come out the other end,” said Indiana University Bloomington biologist Thomas Kaufman, who co-led the project. “One of the things we've learned from this project is that when you compare a lot of different but related genomes, you are more likely to see the genes that are buried in all that A-C-T-G mush.”
Two papers in this week's Nature separately report the results of the four-year genome project and use the data to draw some conclusions about the fruit fly genus Drosophila, particularly its star species, the human nuisance Drosophila melanogaster. Among the papers' conclusions is the idea that resolving any individual species' genome is greatly enhanced when related genomes are compared to it. The project was primarily funded by the National Human Genome Research Institute (NHGRI), part of the National Institutes of Health.
More than 40 “companion” manuscripts are being published or are in press, each of which examines a different aspect of the data produced by the Drosophila 12 Genomes Consortium.
“This remarkable scientific achievement underscores the value of sequencing and comparing many closely-related species, especially those with great potential to enhance our understanding of fundamental biological processes,” said Francis S. Collins, director of NHGRI. “Thanks to the consortium's hard work, scientists around the world now have a rich new source of genomic data that can be mined in many different ways and applied to other important model systems as well as humans.”
The consortium purposely chose a wide variety of fruit flies for study, guessing correctly that both gene similarities and differences among the 12 species would be easier to identify. Some of the Drosophila species the scientists studied are closely related to D. melanogaster, some not. Some of the flies fulfill very specialized ecological niches, such as D. sechellia, which has evolved a unique ability to detoxify the fruit of the Seychelles' noni tree. The other 10 species the consortium examined were D. pseudoobscura, D. simulans, D. yakuba, D. erecta, D. ananassae, D. persimilis, D. willistoni, D. virilis, D. grimshawi, and the cactus-loving D. mojavensis. D. melanogaster's genome was published in 2000 and D. pseudoobscura's genome was published in 2005. The other genomes are newly published.
In comparing the 12 genomes, the scientists found 1,193 new protein-coding genes and hundreds of new functional elements, including regulatory sequences that determine how quickly genes are expressed, and genes that encode functional RNAs such as small nuclear RNAs. They also learned certain genes appear to be evolving faster than others, such as the genes associated with smell and taste, sex and reproduction, and defenses against pathogens.
The Drosophila 12 Genomes Consortium found that D. melanogaster shares about 77 percent of its genes with the other 11 species they studied. The scientists also found errors in about 3 percent of previously sequenced D. melanogaster protein-coding genes, correcting 414 gene sequences on record.
A vexing problem for genomicists is finding genes and other important DNA sequences in heterochromatin, tightly packed areas of chromosomes presumed to experience little expression. Heterochromatin is common in animal genomes.
“The heterochromatin is very hard to analyze,” Kaufman said. “Studies show heterochromatin changes the most. It's full of intermediate- and full-repeat sequences. And there are genes buried in this stuff.”
The conventions for locating the genes that encode proteins are pretty well established. The lingering problem for genomics biologists is locating genes whose parts are interrupted repeatedly, as well as locating genes that do not code for proteins.
By comparing a huge number of genomes, these sorts of genes are relatively easy to locate. Genes that do important things for cells or tissues are more likely to be “conserved” over time; that is, they don't change much despite millions of years of mutations.
One of the companion pieces accompanying this week's Nature papers was written by IUB computational biologist Matthew Hahn. Hahn reports in PLoS Genetics that although all 12 Drosophila species have about the same number of genes (14,000), the genomes are more dynamic than one might expect.
“The highest turnover in gene number occurs in genes involved in sex and reproduction,” Hahn said. “Our results demonstrate that the apparent stasis in total gene number among species has masked rapid turnover in individual gene gain and loss. It is likely that this evolutionary revolving door has played a large role in shaping the morphological, physiological, and metabolic differences among species. This is the reason the 12 species only share 77 percent of their genes.”
Media Contact
More Information:
http://www.indiana.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
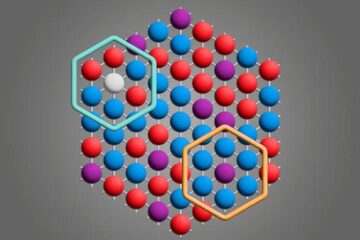
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…