Scientists expand microbe 'gene language'

The Plant-Associated Microbe Gene Ontology (PAMGO) consortium and the GO consortium staff at the European Bioinformatics Institute approved and released more than 450 new terms for describing gene products involved in microbe-host interactions.
The National Science Foundation (NSF) and the USDA's Cooperative State Research, Education and Extension Service (CSREES) support the PAMGO project through grants from their joint Microbial Genome Sequencing program.
This new “common terminology” will speed development of new technologies for preventing infections by disease-causing microbes, while preserving or encouraging the presence of beneficial microbes. Scientists say the Gene Ontology will provide a powerful tool for comparing the functions of genes and proteins in a wide range of disease-related organisms.
“A common set of terms for exchange of information about microbe-host interactions will help researchers communicate information, and expand concepts from studies of microbes and their hosts,” says Maryanna Henkart, director of NSF's Division of Molecular and Cellular Biology.
Microbes that associate with plants or animals can be pathogenic, neutral, or beneficial, but all share many common processes in their interactions with their hosts. For example, all must initially attach to the host. PAMGO, from the beginning, tailored the new terms so they would be useful for describing both benign and pathogenic microbes in plant or animal hosts.
“Having a common set of terms to describe genes of pathogenic and beneficial microbes, as well as the organisms they come into contact with, is critical to understanding host-microbe-environment interactions,” said Brett Tyler, PAMGO project leader at the Virginia Bioinformatics Institute (VBI) in Blacksburg, Va.
Many of the early GO terms described biological functions and processes found in microbes, but very few described the functions used by microbes in their associations with hosts. PAMGO started by creating terms to describe how microbes interact with plants, but researchers soon discovered that almost all the terms were also relevant to microbes that interact with animals and humans.
The Gene Ontology Consortium was organized so all users can actively contribute to the ongoing refinement of the terms. When scientists submit new terms to PAMGO, the entire community participates to synthesize a common understanding of how microbes associate with hosts.
The PAMGO consortium is a collaboration between VBI, Cornell University, North Carolina State University, the University of Wisconsin-Madison, The Institute for Genomic Research (a division of the J.C. Venter Institute) and Wells College. The group works closely with the Gene Ontology Consortium.
PAMGO is working on gene ontology terms for host-association functions of the bacterial pathogens Erwinia chrysanthemi, Pseudomonas syringae pv tomato and Agrobacterium tumefaciens, the fungus Magnaporthe grisea, the oomycetes Phytophthora sojae and Phytophthora ramorum, and the nematode Meloidogyne hapla.
Media Contact
More Information:
http://www.nsf.govAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Attosecond core-level spectroscopy reveals real-time molecular dynamics
Chemical reactions are complex mechanisms. Many different dynamical processes are involved, affecting both the electrons and the nucleus of the present atoms. Very often the strongly coupled electron and nuclear…
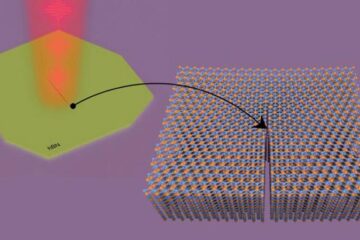
Columbia researchers “unzip” 2D materials with lasers
The new technique can modify the nanostructure of bulk and 2D crystals without a cleanroom or expensive etching equipment. In a new paper published on May 1 in the journal…
Decoding development: mRNA’s role in embryo formation
A new study at Hebrew University reveals insights into mRNA regulation during embryonic development. The study combines single-cell RNA-Seq and metabolic labeling in zebrafish embryos, distinguishing between newly-transcribed and pre-existing…