Common ’signature’ found for different cancers

Discovery yields hope for universal treatment
Researchers at the University of Michigan, Johns Hopkins and the Institute of Bioinformatics in India have discovered a gene-expression “signature” common to distinct types of cancer, renewing hope that a universal treatment for the nation’s second leading killer might be found.
Scientists essentially abandoned the search for a common approach to cancer therapy after research launched by the 1970s “War on Cancer” revealed the many varieties of cancer and the differences among even the same type of cancer in different people. As a result of these discoveries, the focus largely has been on tailoring treatments to specific forms of cancers and even to the precise biology of cancer in a particular person.
“Perhaps we’d learned so much about the differences among cancers that we stopped looking for the similarities. Not having the right tools to look for similarities on a global level didn’t help, either,” says Akhilesh Pandey, assistant professor of biological chemistry in the McKusick-Nathans Institute of Genetic Medicine at Johns Hopkins and chief scientific advisor and founder of the Institute of Bioinformatics, a nonprofit institute located in Bangalore, India.
In the team’s hunt for an overall genetic signature of cancer, which could be useful for diagnosis as well as for developing therapies, the scientists mined a mind-boggling amount of raw information by first creating an online searchable database of 40 published data sets that had collectively analyzed the gene expression “fingerprints” of more than 3,700 cancer tissue samples.
Searching the collected data for common patterns of altered gene expression, the researchers uncovered a “signature” common to all cancers and another that distinguished some kinds of aggressive tumors from their less aggressive counterparts. Their report appears in the June 22 issue of the Proceedings of the National Academy of Sciences.
The signature consisted of 67 genes that were abnormally expressed in all cancers. These genes largely are involved in the cell’s preparation for division — called the cell cycle — and cell proliferation, the researchers report. Since cancers are characterized by uncontrolled cell division, the discovery is logical, even though it wasn’t easy, says Pandey.
“A lot of the available data on gene expression in cancers was just ’warehoused’ — it was there, but not connected to anything,” he says. “We took that data, analyzed it and connected it to relevant information. Now it’s both available and useful.”
Pandey and staff at the Institute of Bioinformatics last year reported creation of the Human Protein Reference Database, an online, searchable, information-rich database of known human proteins and their interactions.
The new project, initiated by Arul Chinnaiyan, M.D., Ph.D., at Michigan, took a similar approach to the cancer problem by developing a way to statistically analyze microarray data and applying the new approach to data from microarray experiments on tumor samples.
Microarray experiments let researchers determine the expression of tens of thousands of genes all at once, providing a molecular “fingerprint” of the tissue sample. Scientists then compare the fingerprint of one sample to that of another — a prostate tumor to normal prostate, or aggressive breast cancer to non-aggressive breast cancer — to identify genes whose expression is higher or lower than “normal.” The idea is that those genes may contribute to the two tissues’ differences.
The mounds of data these experiments create — each identifying hundreds of gene candidates — can be difficult to sift through. But for Chinnaiyan and the research team, the ease with which the data is created meant that a wealth of information about cancers’ genetic profiles already existed, although not in a single form or place.
Answering some critics who claim that experimental differences make microarray data virtually impossible to compare, Pandey says that the difficulty actually supports their results. “If some people consider these sets to be so different as to be incomparable, then anything that does turn out to be common to all of them seems pretty likely to be real,” he suggests.
The researchers also validated their proposed cancer signature by examining data sets published after creation of the database, dubbed ONCOMINE. The same signature discriminated between cancer and normal tissue in seven of nine new data sets, including properly discriminating three types of cancer not used to create the database, the scientists report.
ONCOMINE connects the cancer microarray database to several sources of additional information, including the scientific literature, the Human Protein Reference Database and Online Inheritance in Man, the online catalog of all proven disease-gene connections. ONCOMINE is owned by the University of Michigan, and is available online to academic researchers free-of-charge following registration.
Authors on the report are Daniel Rhodes, Jianjun Yu, Radhika Varambally, Debashis Ghosh, Terrence Barrette and Chinnaiyan of the University of Michigan Medical School; Kalyan Shanker and Nandan Deshpande of the Institute of Bioinformatics; and Pandey of Johns Hopkins. Pandey does not receive compensation for his role as scientific adviser to the Institute of Bioinformatics.
Media Contact
More Information:
http://www.jhmi.eduAll latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

Why getting in touch with our ‘gerbil brain’ could help machines listen better
Macquarie University researchers have debunked a 75-year-old theory about how humans determine where sounds are coming from, and it could unlock the secret to creating a next generation of more…

Attosecond core-level spectroscopy reveals real-time molecular dynamics
Chemical reactions are complex mechanisms. Many different dynamical processes are involved, affecting both the electrons and the nucleus of the present atoms. Very often the strongly coupled electron and nuclear…
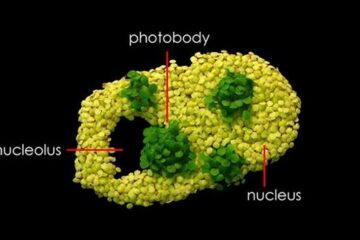
Free-forming organelles help plants adapt to climate change
Scientists uncover how plants “see” shades of light, temperature. Plants’ ability to sense light and temperature, and their ability to adapt to climate change, hinges on free-forming structures in their…