Untangling Whole Genomes of Individual Species From a Microbial Mix

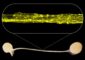
Joshua N. Burton, University of Washington Hi-C data can cluster contigs from a metagenome assembly into species. Each of the 12 species with a substantial presence in the draft assembly is represented by a cluster. Each contig is shown as a dot, with size indicating contig length, colored by species. Edge widths represent the densities of Hi-C links between the contigs shown. Only 2400 contigs are shown: the 200 largest contigs that map uniquely to each species.
A new approach to studying microbes in the wild will allow scientists to sequence the genomes of individual species from complex mixtures. It marks a big advance for understanding the enormous diversity of microbial communities —including the human microbiome. The work is described in an article published May 22 in Early Online form in the journal G3: Genes|Genomes|Genetics, published by the Genetics Society of America.
“This new method will allow us to discover many currently unknown microbial species that can’t be grown in the lab, while simultaneously assembling their genome sequences,” says co-author Maitreya Dunham, a biologist at the University of Washington’s Department of Genome Sciences.
Microbial communities, whether sampled from the ocean floor or a human mouth, are made up of many different species living together. Standard methods for sequencing these communities combine the information from all the different types of microbes in the sample. The result is a hodgepodge of genes that is challenging to analyze, and unknown species in the sample are difficult to discover.
“Our approach tells us which sequence fragments in a mixed sample came from the same genome, allowing us to construct whole genome sequences for individual species in the mix,” says co-author Jay Shendure, also of the University of Washington’s Department of Genome Sciences.
The key advance was to combine standard approaches with a method that maps out which fragments of sequence were once near each other inside a cell. The cells in the sample are first treated with a chemical that links together DNA strands that are in close proximity. Only strands that are inside the same cell will be close enough to link. The DNA is then chopped into bits, and the linked portions are isolated and sequenced.
“This elegant method enables the study of microbes in the environment,” says Brenda Andrews, editor-in-chief of the journal G3: Genes|Genomes|Genetics. Andrews is also Director of the Donnelly Centre and the Charles H. Best Chair of Medical Research at the University of Toronto. “It will open many windows into an otherwise invisible world.”
At a time when personal microbiome sequencing is becoming extremely popular, this method breaks important ground in helping researchers to build a complete picture of the genomic content of complex mixtures of microorganisms. This complete picture will be crucial for understanding the impact of varying microbiome populations and the relevance of particular microorganisms for individual health.
CITATION: Species-Level Deconvolution of Metagenome Assemblies with Hi-C-Based Contact Probability Maps Joshua N. Burton, Ivan Liachko, Maitreya J. Dunham, and Jay Shendure. G3: Genes|Genomes|Genetics g3.114.011825; Early Online May 22, 2014, doi:10.1534/g3.114.011825; PMID 24855317.
FUNDING INFORMATION: This work was supported by NIH/NHGRI grant T32HG000035 (J.N.B.), NIH/NHGRI grant HG006283 (J.S.), NIH/NIGMS grant P41 GM103533 (I.L. & M.J.D.), NSF grant 1243710 (I.L. & M.J.D.), DOE/-LBL-JGI grant 7074345/DE-AC02-05CH11231 (J.S.). M.J.D. is a Rita Allen Foundation Scholar and a Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research.
####
ABOUT G3: G3:Genes|Genomes|Genetics publishes high-quality, valuable findings, regardless of perceived impact. G3 publishes research that generates useful genetic and genomic information such as genome maps, single gene studies, QTL studies, mutant screens and advances in methods and technology, novel mutant collections, genome-wide association studies (GWAS) including gene expression, SNP and CNV studies; exome sequences related to a specific disease, personal exome and genome sequencing case, disease and population reports, and more.
Conceived by the Genetics Society of America, with its first issue published June 2011, G3 is fully open access. G3 uses a Creative Commons license that allows the most free use of the data, which anyone can download, analyze, mine and reuse, provided that the authors of the article receive credit. GSA believes that rapid dissemination of useful data is the necessary foundation for analysis that leads to mechanistic insights. It is our hope is that this strategy will spawn new discovery.
ABOUT GSA: Founded in 1931, the Genetics Society of America (GSA) is the professional scientific society for genetics researchers and educators. The Society’s more than 5,000 members worldwide work to deepen our understanding of the living world by advancing the field of genetics, from the molecular to the population level. GSA promotes research and fosters communication through a number of GSA-sponsored conferences including regular meetings that focus on particular model organisms. GSA publishes two peer-reviewed, peer-edited scholarly journals: GENETICS, which has published high quality original research across the breadth of the field since 1916, and G3:Genes|Genomes|Genetics, an open-access journal launched in 2011 to disseminate high quality foundational research in genetics and genomics. The Society also has a deep commitment to education and fostering the next generation of scholars in the field. For more information about GSA, please visit www.genetics-gsa.org. Follow GSA on Facebook at facebook.com/GeneticsGSA and on Twitter @GeneticsGSA.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…