Small molecule acts as on-off switch for nature's antibiotic factory
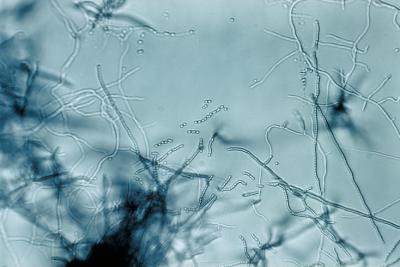
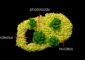
The soil bacteria Streptomyces form filamentous branches that extend into the air to create spiraling towers of spores. Duke researchers have discovered the switch that can turn off sporulation and turn on antibiotic production. Credit: US Centers for Disease Control and Prevention
Scientists have identified the developmental on-off switch for Streptomyces, a group of soil microbes that produce more than two-thirds of the world's naturally derived antibiotic medicines.
Their hope now would be to see whether it is possible to manipulate this switch to make nature's antibiotic factory more efficient.
The study, appearing August 28 in Cell, found that a unique interaction between a small molecule called cyclic-di-GMP and a larger protein called BldD ultimately controls whether a bacterium spends its time in a vegetative state or gets busy making antibiotics.
Researchers found that the small molecule assembles into a sort of molecular glue, connecting two copies of BldD as a cohesive unit that can regulate development in the Gram-positive bacteria Streptomyces.
“For decades, scientists have been wondering what flips the developmental switch in Streptomyces to turn off normal growth and to begin the unusual process of multicellular differentiation in which it generates antibiotics,” said Maria A. Schumacher, Ph.D., an associate professor of biochemistry at the Duke University School of Medicine. “Now we not only know that cyclic-di-GMP is responsible, but we also know exactly how it interacts with the protein BldD to activate its function.”
Streptomyces has a complex life cycle with two distinct phases: the dividing, vegetative phase and a distinct phase in which the bacteria form a network of thread-like filaments to chew up organic debris and churn out antibiotics and other metabolites. At the end of this second phase, the bacteria form filamentous branches that extend into the air to create spiraling towers of spores.
In 1998 researchers discovered a gene that kept cultured Streptomyces bacteria from creating these spiraling towers of fuzz on their surface. They found that this gene, which they named BldD to reflect this “bald” appearance, also affected the production of antibiotics.
Subsequent studies have shown that BldD is a special protein called a transcription factor, a type of master regulator that binds DNA and turns on or off more than a hundred genes to control biological processes like sporulation. But in more than a decade of investigation, no one had been able to identify the brains behind the operation, the molecule that ultimately controls this master regulator in Streptomyces.
Then scientists at the John Innes Centre in the United Kingdom — where much of the research on Streptomyces began — discovered that the small molecule cyclic-di-GMP is generated by several transcription factors regulated by BldD. The researchers did a quick test to see if this small molecule would itself bind BldD, and were amazed to find that it did. They contacted longtime collaborators Schumacher and Richard G. Brennan Ph.D. at Duke to see if they could take a closer look at this important interaction.
The Duke team used a tool known as x-ray crystallography to create an atomic-level three-dimensional structure of the BldD-(cyclic-di-GMP) complex.
BldD normally exists as a single molecule or monomer, but when it is time to bind DNA and suppress sporulation, it teams up with another copy of itself to do the job. The 3D structure built by the researchers revealed that these two copies of BldD never physically touch, and instead are stuck together by four copies of cyclic-di-GMP.
“We have looked through the protein databank and scoured our memories, but this finding appears to be unique,” said Brennan, who is a professor and chair of biochemistry at Duke University School of Medicine. “We have never seen a type of structure before where two monomers become a functional dimer, with no direct interaction between them except a kind of small-molecule glue.”
To confirm their findings, Schumacher determined several crystal structures from different flavors of bacteria (S. venezuelae and S. coelicolor) and came up with the same unusual result every time.
Now that the researchers know how cyclic-di-GMP and BldD can become glued together to turn off sporulation and turn on antibiotic production, they would like to know how the complex can become unglued again to flip the switch the other way.
The research was supported by a Long Term EMBO Fellowship (ALTF 693-2012), a Leopoldina Postdoctoral Fellowship, the Biotechnology and Biological Sciences Research Council (BB/H006125/1), the MET Institute Strategic Programme, and the Duke University School of Medicine.
CITATION: “Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development,” Natalia Tschowri, Maria A. Schumacher, Susan Schlimpert, Nagababu Chinnam, Kim C. Findlay, Richard G. Brennan, and Mark J. Buttner. Cell, August 28, 2014.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…