New science of metagenomics 'will transform modern microbiology'

Microorganisms are essential to life on Earth, transforming key elements into energy, maintaining the chemical balance in the atmosphere, providing plants and animals with nutrients, and performing other functions necessary for survival. There are billions of benign microbes in the human body, for example, that help to digest food, break down toxins, and fight off disease-causing microbes. Microbes are used commercially for many purposes, including making antibiotics, remediating oil spills, enhancing crop production, and producing biofuels.
Historically, microbiology focused on the study of individual species of organisms that could be grown in a laboratory and examined under a microscope, but most of the life-supporting activities of microbes are carried out by complex communities of microorganisms, and many cannot be grown in laboratory culture. Metagenomics will transform modern microbiology by giving scientists the tools to study entire communities of microbes — the vast majority of which are likely to be previously unknown species that cannot be cultured — and how they interact to perform such functions as balancing the atmosphere's composition, fighting disease, and supporting plant growth, the new report says.
“Metagenomics lets us see into the previously invisible microbial world, opening a frontier of science that was unimaginable until recently,” said Jo Handelsman, Howard Hughes Medical Institute Professor, departments of plant pathology and bacteriology, University of Wisconsin, Madison, and co-chair of the committee that wrote the report. “Shedding light on thousands of new microorganisms will lead to new biological concepts as well as practical applications for human health, agriculture, and environmental stewardship.”
Metagenomics studies begin by extracting DNA from all the microbes living in a particular environmental sample; there could be thousands or even millions of organisms in one sample. The extracted genetic material consists of millions of random fragments of DNA that can be cloned into a form capable of being maintained in laboratory bacteria. These bacteria are used to create a “library” that includes the genomes of all the microbes found in a habitat, the natural environment of the organisms. Although the genomes are fragmented, new DNA sequencing technology and more powerful computers are allowing scientists to begin making sense of these metagenomic jigsaw puzzles. They can examine gene sequences from thousands of previously unknown microorganisms, or induce the bacteria to express proteins that are screened for capabilities such as vitamin production or antibiotic resistance.
The Research Council report was requested by several federal agencies interested in the potential of metagenomics and how best to encourage its success. In particular, the committee was asked to recommend promising directions for future studies. It concluded that the most efficient way to boost the field of metagenomics overall would be to establish a Global Metagenomics Initiative that includes a few large-scale, internationally coordinated projects and numerous medium- and small-size studies.
“Because the challenges and opportunities presented by metagenomics are so enormous, a major commitment equivalent to that of the Human Genome Project is both justified and necessary,” added committee co-chair James M. Tiedje, University Distinguished Professor of Microbiology and director of the Center for Microbial Ecology, Michigan State University, East Lansing.
The goal of the large projects should be to characterize in great detail carefully chosen microbial communities in habitats worldwide, the report says. These studies could unite scientists from multiple disciplines around the study of a particular sample, habitat, function, or analytical challenge, the report adds. For example, one large project could focus on the microbial community associated with the human body, while others could focus on soil and seawater, or managed environments such as sludge processing sites.
The large projects would be “virtual” centers collecting data from scientists working at many locations around the world, and would probably need to be sustained for 10 years, the report notes. They would also serve as incubators for the development of novel techniques and community databases that would inform investigators running smaller experiments. In addition, the large studies would provide the “big science” appeal that is often useful in igniting public interest.
The metagenomics research community will include scientists funded by many government agencies working on many different habitats. These scientists should be encouraged to work together to disseminate advances, agree on common standards, and develop guidelines for best practices, the committee said. Such collaboration between government agencies should be facilitated by a group like the Microbe Project, an interagency committee that was formed in 2000 to advance “genome-enabled microbial science.”
Noting that the data generated by the Human Genome Project were quickly made available in a public database, making them more valuable to researchers, the committee urged that metagenomic data likewise be made publicly available in international archives as rapidly as possible. The databases should include not only gene sequences but also information about sampling and DNA extraction techniques, as well as the computational and algorithmic methods used to analyze the data.
Media Contact
More Information:
http://www.nas.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
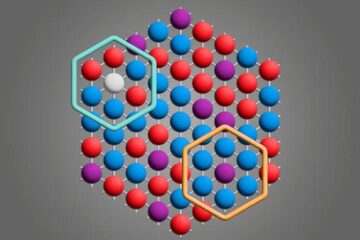
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…