New technique boosts by four times the size of a protein that researchers can analyze

But now, Cornell researchers have extended a powerful technique to increase by fourfold the size of a protein that can be analyzed, to those containing more than 2,000 amino acids, up from about 500.
Called a “top-down” approach, the technique uses a mass spectrometer, which measures the masses of ions or charged particles. Researchers break up the protein into pieces and weigh both the masses of the whole protein and of the individual pieces. By matching the weight of the whole protein and its pieces with those of known protein sequences in a database, they can identify the protein. Any differences in mass with known proteins can help researchers also find where and how proteins have been modified in cells.
For example, if a section of a protein has an increased weight of 16, researchers can tell that the protein has been oxidized within that section, which means that an oxygen atom (with an atomic weight of 16) was added.
“When you isolate a protein from a mixture, your first problem is to know which one it is,” said Fred McLafferty, Cornell's Peter J.W. Debye Professor Emeritus of Chemistry and Chemical Biology and senior author of the paper published in the Oct. 6 issue of the journal Science. “Mass spectrometry characterizes a protein by measuring the masses produced from it.”
The new technique provides a far more efficient way to break down proteins inside the mass spectrometer — sprayed, heated and then bashed with gas molecules — to obtain their pieces for the mass analysis. They are sprayed in an electric field where they pick up charges and are vaporized as they pass through a heated capillary. Then they are hit by air molecules in low-energy collisions to keep the protein from folding up; then they are slammed with air molecules in high-energy collisions to dissociate the protein. Next, the mass spectrometer weighs any surviving protein and its pieces simultaneously.
The top-down approach rivals the commonly used “bottom-up” approach, in which proteins are first broken down, or digested, enzymatically into smaller units of five to 20 amino acids. These pieces are then introduced into the mass spectrometer where researchers can match masses of those pieces to known sequences and identify the proteins.
However, this approach reveals less information concerning modifications to proteins. Such important cellular processes as oxidation and acetylation of proteins add chemical groups and alter how a protein functions, such as modifying an enzymatic pathway. The bottom-up approach is limited because the samples seldom have masses that represent all the pieces of the protein, and masses can usually be matched to several combinations of pieces and modifications. Therefore, the method rarely locates all modifications in newly isolated proteins.
“Each approach has different strengths and weaknesses,” said Mi Jin, a postdoctoral associate in chemistry and chemical biology and one of the paper's lead authors with fellow graduate student Xuemei Han. Jin said that the bottom-up approach is often used for larger scale studies of proteins because it can identify a large number of proteins from a sample, but it does not provide a complete picture of each protein. The top-down approach, on the other hand, measures the whole protein and thus provides more confident identifications; it is also better at revealing modifications and mutations, where there might be a mistake or addition in the sequence. The new approach can also provide sequence information on a protein from scratch when it is not present in any database.
The study was funded by National Institutes of Health.
Media Contact
More Information:
http://www.cornell.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
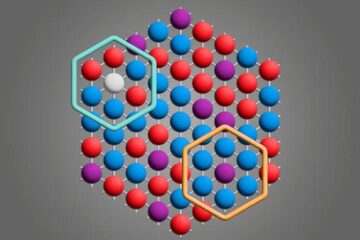
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…