Research pinpoints West Nile virus antibody binding site

“Science doesn't yet fully understand exactly how neutralizing antibodies work,” said Michael Rossmann, the Hanley Distinguished Professor of Biological Sciences in Purdue's College of Science. “This work has shown precisely where the antibody binds to the virus, and we now have a theory for how it interacts with the virus to disarm it. Perhaps we are starting to understand why this particular antibody can inhibit the infectivity of the virus, which is important to understand if a vaccine is going to be developed.”
Purdue worked with researchers from the Washington University School of Medicine in St. Louis.
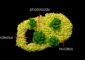
West Nile belongs to a family of viruses known as flaviviruses, which includes a number of dangerous insect-borne disease-causing viruses. The antibody attaches to a protein called an E protein, for envelope protein, which makes up the virus's outer shell. There are 180 copies of E proteins symmetrically arranged in 60 sets of three, forming a geometric shape called an icosahedron, which is made up of triangular facets.
The researchers, however, were surprised to discover that this antibody recognizes only two of the E proteins in each set of three, said Bärbel Kaufmann, a postdoctoral research associate working in the Rossmann lab.
“This finding was very unusual,” she said. “If the E proteins really are the same, why doesn't the antibody bind to all of the E proteins? This kind of asymmetry, where you have two proteins binding and one not binding, has not been seen before.”
The researchers theorize that, although chemically identical, these E proteins exist in different environments relative to each other and might, therefore, have slightly different structures, said Richard J. Kuhn, a professor and head of Purdue's Department of Biological Sciences.
The findings were detailed in a research paper appearing last week in Proceedings of the National Academy of Sciences.
Researchers know that when the virus infects a host cell, it interacts with the cell membrane in such a way that it is swallowed up by the membrane and enters the cell as an “endocytotic particle.” The Purdue researchers have now developed a theory for the mechanism behind the interaction and will test it in further research, Rossmann said.
Once the virus penetrates the host cell, the viral membrane fuses with an internal membrane in the cell. This process causes the virus particle to empty its contents inside the cell and leads to infection.
To study how the antibodies and E proteins attach, Kaufmann first separated the antibody's tail end from its two grasping, fingerlike structures called “antigen binding fragments.”
“We don't want to handle the whole molecule, so we cut off these antigen binding domains and then combined them with the virus, forming the virus-antibody complex,” Kaufmann said.
The researchers then used an electron microscope and a process called cryoelectron microscopy to take detailed pictures of this complex. Then they computed a three-dimensional model based on these pictures showing the outstretched antigen binding fragments attached to the virus particle.
The E proteins in one triangular segment of the icosahedron are not all positioned the same way relative to each other and to the various “axes” that define the icosahedron. This difference in position appears to be crucial in the binding process.
The antibody is called a monoclonal antibody because it recognizes only a single binding site on the E protein. Each E protein has three “domains,” or well-defined, folded segments. The third domain has a structure commonly seen in molecules that attach to proteins to perform a specific function in cells. The grasping antibody segment used in the study attaches only to the third domain of the E proteins.
“One of the three E proteins in the triangular segment fails to be recognized by the antibody because its third domain crowds together with other E proteins on the viral surface,” Kuhn said.
Researchers at the Washington University School of Medicine have shown through experiments that the antibody does not prevent the virus from attaching to human cells, so it is likely that the antibody works by preventing the fusion step from occurring.
Purdue researchers had previously theorized that before the virus fuses, the E protein undergoes dramatic structural repositioning and that the antibodies might inhibit those changes, preventing infection.
“We have a model for understanding how this inhibition process may work,” Rossmann said. “We think it's likely that the antibody blocks the positional changes needed for the E protein before fusion, in effect preventing the virus from infecting the cell by jamming the mechanism.”
After infection, it takes a few days to a few weeks for the body to make the antibodies, or enough of them to fend off infection.
“If the virus is fast enough, you become sick,” Kuhn said. “But if you were vaccinated, you might already have enough antibodies to prevent infection, and that's why these findings are ultimately important. If we understand the mechanism of neutralization, then we might be able to design more effective vaccines.”
West Nile virus causes a potentially fatal illness and has infected thousands of people in the United States over the past five years, killing more than 700 people in that time frame.
The research is funded by the National Institutes of Health. Future work could focus on confirming the theoretical model for the antibody's neutralizing mechanism.
“If our theory is right and the antibody binding blocks the E protein's transformation so that fusion can't take place, then we should be able to capture the intermediate stages of this rearrangement of the E protein prior to fusion,” Kaufmann said.
The paper was authored by Kaufmann, Purdue electron microscopist Paul R. Chipman, Purdue associate research scientist Wei Zhang, Kuhn and Rossmann, and three researchers from the Washington University School of Medicine: Grant E. Nybakken, a graduate student in the Department of Pathology and Immunology; Michael S. Diamond, an assistant professor in the departments of Medicine, Molecular Microbiology, Pathology and Immunology; and Daved H. Fremont, an associate professor of pathology and immunology, biochemistry and molecular physics.
Writer: Emil Venere, (765) 494-4709, venere@purdue.edu
Sources: Michael Rossmann, (765) 494-4911, mgr@indiana.bio.purdue.edu
Richard J. Kuhn, (765) 494-1164, kuhnr@purdue.edu
Barbel Kaufmann, bkaufman@bilbo.bio.purdue.edu
Purdue News Service: (765) 494-2096; purduenews@purdue.edu
Media Contact
More Information:
http://www.purdue.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Microscopic basis of a new form of quantum magnetism
Not all magnets are the same. When we think of magnetism, we often think of magnets that stick to a refrigerator’s door. For these types of magnets, the electronic interactions…

An epigenome editing toolkit to dissect the mechanisms of gene regulation
A study from the Hackett group at EMBL Rome led to the development of a powerful epigenetic editing technology, which unlocks the ability to precisely program chromatin modifications. Understanding how…

NASA selects UF mission to better track the Earth’s water and ice
NASA has selected a team of University of Florida aerospace engineers to pursue a groundbreaking $12 million mission aimed at improving the way we track changes in Earth’s structures, such…