Lineage trees for cells

Weizmann Institute scientists develop new analytical method
Some fundamental outstanding questions in science – “Where do stem cells originate?” “How does cancer develop?” “When do cell types split off from each other in the embryo?” – might be answered if scientists had a way to map the history of the body’s cells going back to the fertilized egg. Now, a multidisciplinary team at the Weizmann Institute of Science has developed an analytical method that can trace the lineage trees of cells.
This accomplishment started with a challenge to common wisdom, which says that every cell in an organism carries an exact duplicate of its genome. Although mistakes in copying, which are passed on to the next generation of cells as mutations, occur when cells divide, such tiny flaws in the genome are thought to be trivial and mainly irrelevant. But research students Dan Frumkin and Adam Wasserstrom of the Institute’s Biological Chemistry Department, working under the guidance of Prof. Ehud Shapiro of the Biological Chemistry and Computer Science and Applied Mathematics Departments, raised a new possibility: though biologically insignificant, the accumulated mutations might hold a record of the history of cell divisions. These findings were published today in PLoS Computational Biology.
Together with Prof. Uriel Feige of the Computer Science and Applied Mathematics Department and research student Shai Kaplan, they proved that these mutations can be treated as information and used to trace lineage on a large scale, and then applied the theory to extracting data and drafting lineage trees for living cells.
Methods employed until now for charting cell lineage have relied on direct observation of developing embryos. This method worked well enough for the tiny, transparent worm, C. elegans, which has a total of about 1,000 cells, but for humans, with 100 trillion cells, or even newborn mice or human embryos at one month, each of which has one billion cells after some 40 rounds of cell division, the task would be impossible.
The study focused on mutations in specific mutation-prone areas of the genome known as microsatellites. In microsatellites, a genetic “phrase” consisting of a few nucleotides (genetic “letters”) is repeated over and over; mutations manifest themselves as additions or subtractions in length. Based on the current understanding of the mutation process in these segments, the scientists proved mathematically that microsatellites alone contain enough information to accurately plot the lineage tree for a one-billion-cell organism.
Both human and mouse genomes contain around 1.5 million microsatellites, but the team’s findings demonstrated that a useful analysis can be performed based on a much smaller number. To obtain a consistent mutation record, the team used organisms with a rare genetic defect found in plants and animals alike. While healthy cells have repair mechanisms to correct copying mistakes and prevent mutation, cells with the defect lack this ability, allowing mutations to accumulate relatively rapidly.
Borrowing a computer algorithm used by evolutionary biologists that analyzes genetic information in order to place organisms on branches of the evolutionary tree, the researchers assembled an automated system that samples the genetic material from a number of cells, compares it for specific mutations, applies the algorithm to assess degrees of relatedness, and then outlines the cell lineage tree. To check their system, they pitted it against the tried-and-true method of observing cell divisions as they occurred in a lab-grown cell culture. The team found that, from an analysis of just 50 microsatellites, they could successfully recreate an accurate cell lineage tree.
While the research team plans to continue to test their system on more complex organisms such as mice, several scientists have already expressed interest in integrating the method into ongoing research in their fields. Says Prof. Shapiro, who heads the project: “Our discovery may point the way to a future ’Human Cell Lineage Project’ that would aim to resolve fundamental open questions in biology and medicine by reconstructing ever larger portions of the human cell lineage tree.”
Media Contact
More Information:
http://www.weizmann.ac.il/udi/plos2005All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
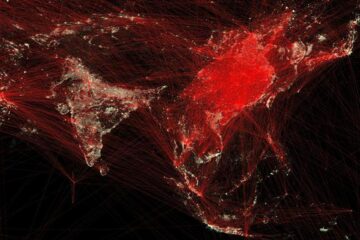
Economies take off with new airports
A global study by an SUTD researcher in collaboration with scientists from Japan explores the economic benefits of airport investment in emerging economies using nighttime satellite imagery. Be it for…
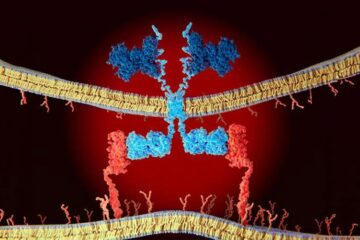
CAR T–cell immunotherapy targets
Pan-cancer analysis uncovers a new class of promising CAR T–cell immunotherapy targets. Scientists at St. Jude Children’s Research Hospital found 156 potential CAR targets across the brain and solid tumors,…

Stony coral tissue loss disease
… is shifting the ecological balance of Caribbean reefs. The outbreak of a deadly disease called stony coral tissue loss disease is destroying susceptible species of coral in the Caribbean…





















