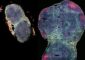
New bioinformatics tool to visualize transcriptomes

Next-generation sequencing has revolutionized functional genomics, with protocols such as RNA-seq, ChIP-seq and CAGE being used widely around the world.
The power of these techniques lies in the fact that they enable the genome-wide discovery of transcripts and transcription factor binding sites, which is key to understanding the molecular mechanisms underlying cell function in healthy and diseased individuals and the development of diseases like cancer.
The integration of data from multiple experiments is an important aspect of the interpretation of results, however the growing number of datasets generated makes a thorough comparison and analysis of results cumbersome.
In a report published today in the journal Nature Biotechnology, Jessica Severin and colleagues describe the development of ZENBU, a tool that combines a genome browser with data analysis and a linked expression view, to facilitate the interactive visualization and comparison of results from large numbers of next-generation sequencing datasets.
The key difference between ZENBU and previous tools is the ability to dynamically combine thousands of experimental datasets in an interactive visualization environment through linked genome location and expression signal views.
This allows scientists to compare their own experiments against the over 6000 ENCODE and FANTOM consortium datasets currently loaded into the system, thus enabling them to discover new and interesting biological mechanisms. The tool is designed to integrate millions of experiments/datasets of any kind (RNA-seq, ChIP-seq or CAGE), hence its name: zenbu means 'all' or 'everything' in Japanese.
ZENBU is freely available for use on the web and for installation in individual laboratories, and all ZENBU sites are connected and continuously share data. The tool can be accessed or downloaded from http://fantom.gsc.riken.jp/zenbu/.
“By distributing the data and servers we encourage scientists to load and share their published data to help build a comprehensive resource to further advance research efforts and collaborations around the world,” explain the authors.
ZENBU is accessible at http://fantom.gsc.riken.jp/zenbu/
Dr. Forrest and Dr. Severin are available for interviews in English by email. Please contact:
Alistair Forrest, Team leader
Genome Information Analysis Team
Life Science Accelerator Technology Group
Division of Genomic Technologies
RIKEN Center for Life Science Technologies
Email: forrest@gsc.riken.jp
Jessica Severin, Senior Technical Scientist
Genome Information Analysis Team
Life Science Accelerator Technology Group
Division of Genomic Technologies
RIKEN Center for Life Science Technologies
Email: severin@gsc.riken.jp
Alternatively, for more information please contact:
Juliette Savin
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225
Email: pr@riken.jp
A copy of the original Nature Biotechnology article and a figure are available on request.
Reference
- Jessica Severin, Marina Lizio, Jayson Harshbarger, Hideya Kawaji, Carsten O Daub, Yoshihide Hayashizaki, the FANTOM consortium, Nicolas Bertin, and Alistair RR Forrest. “Interactive visualization and analysis of large-scale NGS data-sets using ZENBU”. Nature Biotechnology, http://dx.doi.org/10.1038/nbt.2840 (2013)
About RIKEN
RIKEN is Japan's flagship research institute for basic and applied research. Over 2500 papers by RIKEN researchers are published every year in reputable scientific and technical journals, covering topics ranging across a broad spectrum of disciplines including physics, chemistry, biology, medical science and engineering. RIKEN's advanced research environment and strong emphasis on interdisciplinary collaboration has earned itself an unparalleled reputation for scientific excellence in Japan and around the world.
Website: http://www.riken.jp Find us on Twitter at @riken_en
About the Center for Life Science Technologies (CLST) The RIKEN Center for Life Science Technologies aims at the development of key technologies for breakthroughs in medical and pharmaceutical applications by conducting ground-breaking research and development programs for next-generation life sciences. CLST comprises the Division of Structural and Synthetic Biology, the Division of Genomic Technologies, and the Division of Bio-function Dynamics Imaging, which will work together in this endeavor. Research and development programs are carried out in collaboration with companies, universities, and international consortia, in order to disseminate the center's achievements to the global community.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Red light therapy for repairing spinal cord injury passes milestone
Patients with spinal cord injury (SCI) could benefit from a future treatment to repair nerve connections using red and near-infrared light. The method, invented by scientists at the University of…
Insect research is revolutionized by technology
New technologies can revolutionise insect research and environmental monitoring. By using DNA, images, sounds and flight patterns analysed by AI, it’s possible to gain new insights into the world of…

X-ray satellite XMM-newton sees ‘space clover’ in a new light
Astronomers have discovered enormous circular radio features of unknown origin around some galaxies. Now, new observations of one dubbed the Cloverleaf suggest it was created by clashing groups of galaxies….