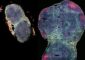
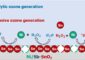
Marking metabolism

Recent advances in DNA sequencing have made it relatively easy to acquire the full genotype of an individual, but it is equally important to match those genes to their functions. One useful step is to build up a ‘metabolic phenotype’ outlining all the processes operating to sustain the individual’s life.
Jun Kikuchi and co-workers at the RIKEN Plant Science Center in Yokohama, Yokohama City University and Nagoya University have developed a systematic method to characterize metabolic pathways in plants and animals. Their method involves measuring nuclear magnetic resonance (NMR) of samples and comparing them against an extensive database of molecules associated with metabolism, known as metabolites.
NMR works by detecting the response of atoms or molecules to a magnetic field. Normal carbon atoms show no response, so cells must be labeled with the stable isotope carbon-13.
Kikuchi and co-workers fed Arabidopsis plants and silkworm larvae with glucose and amino acids that had carbon-13 atoms in place of the normal carbon. After this incubation process, almost all the metabolites produced by the cells contained carbon-13. Importantly, carbon-13 displays a slightly different magnetic response depending on the structure of the molecule it is in, so each metabolite provided a unique NMR spectrum.
The researchers compared the spectra of their samples against a database of spectra for known metabolites. They identified 57 unique metabolites in the silkworm larvae, and 61 in Arabidopsis.
The team then used a technique called Principal Component Analysis to identify correlations between metabolites in the silkworm. These correlations represent metabolic pathways related to key stages in the larval development.
In particular, the results showed a random pattern of metabolic pathways over the first six days of the study, giving way to some correlations later. This suggests that better metabolic organization emerged as the larvae grew.
The study represents the first ‘top-down’ method of analyzing whole metabolic pathways. It provides a macroscopic phenotype describing cells, fluids and tissues, rather than looking at specific reactions from the atomic level upwards. What’s more, the technique is relatively quick.
“After an NMR measurement, typically taking about 1 hour, computation of the metabolic pathways finishes within half a day,” explains Eisuke Chikayama, who wrote the team’s recent paper in PLoS ONE (1).
Chikayama is also hopeful that the technique could be extended to other plants and animals, including humans.
“Our method is not restricted to any particular organism, if adequate NMR samples are ready.”
Reference
1. Chikayama, E., Suto, M., Nishihara, T., Shinozaki, K., Hirayama, T. & Kikuchi J. Systematic NMR analysis of stable isotope labeled metabolite mixtures in plant and animal systems: Coarse grained views of metabolic pathways. PLoS ONE 3(11), e3805 (2008).
The corresponding author for this highlight is based at the RIKEN Metabolomics Research Group: Advanced NMR Metabomics Research Unit
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Red light therapy for repairing spinal cord injury passes milestone
Patients with spinal cord injury (SCI) could benefit from a future treatment to repair nerve connections using red and near-infrared light. The method, invented by scientists at the University of…
Insect research is revolutionized by technology
New technologies can revolutionise insect research and environmental monitoring. By using DNA, images, sounds and flight patterns analysed by AI, it’s possible to gain new insights into the world of…

X-ray satellite XMM-newton sees ‘space clover’ in a new light
Astronomers have discovered enormous circular radio features of unknown origin around some galaxies. Now, new observations of one dubbed the Cloverleaf suggest it was created by clashing groups of galaxies….