How a microbial biorefinery regulates genes
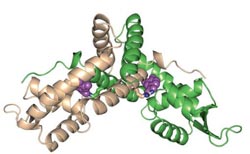
The protein PcaV in the presence of protocatechuate<br><br> Digesting lignin, a very stable, plentiful polymer in biomass, is a limiting step in the production of biofuels. Brown researchers have identified a microscopic chemical switch that lets bacteria get to work, breaking lignin down into its component parts. <br><br>Credit: Sello lab/Brown University <br>
Researchers at Brown have figured out the microscopic chemical switch that allows Streptomyces bacteria to get to work, breaking lignin down into its constituent parts.
Microorganisms that can break down plant biomass into the precursors of biodiesel or other commodity chemicals might one day be used to produce alternatives to petroleum. But the potential of this “biorefinery” technology is limited by the fact that most microorganisms cannot break down lignin, a highly stable polymer that makes up as much as a third of plant biomass.
Streptomyces bacteria are among few microorganisms known to degrade and consume lignin. Now a group of researchers at Brown University has unlocked the genetic and molecular mechanisms behind a key part of that process. The results are published in the journal Nucleic Acids Research.
Jason Sello, professor of chemistry at Brown, and Rebecca Page, professor in biology in the Department of Molecular Biology, Cell Biology, and Biochemistry, directed the research with graduate students Jennifer Davis and Breann Brown.
“Aside from the implications for biotechnology, this work is significant because it yielded fundamental insights into how bacteria control the expression of their genes,” Sello said. “Understanding how genes underlying lignin degradation are regulated could have practical importance in that we could possibly use this information to engineer bacteria that can convert this important component of plant biomass into the biofuels of high-value chemicals.”
The consumption of lignin by Streptomyces bacteria is a multistep process. First, the bacteria release enzymes that depolymerize the lignin — break it down into its constituent compounds. The bacteria take up the resulting compounds and use the carbon to support their growth and reproduction. Some of that lignin-derived carbon is converted into triglycerides, the building blocks of biodiesel, and other high-value compounds.
Seed money for a promising idea
Jason Sello and Rebecca Page received a University seed grant to test their model of genetic and molecular mechanisms behind the digestion of lignin by Streptomyces bacteria. Credit: Mike Cohea/Brown UniversityThis latest research deals with the second part of that process, the ability of Streptomyces bacteria to metabolize aromatic compounds derived from lignin, which are highly stable and hard to break down.
In previous work, Sello and Davis identified a cluster of genes in Streptomyces bacteria that encode enzymes for breaking down a lignin-derived compound called protocatechuate. Under normal circumstances, those genes were inactive — essentially switched off. Only when bacteria were grown in a medium containing protocatechuate did the genes switch on and produce the appropriate enzymes. In an effort to understand this phenomenon, Sello and Davis discovered that a transcription factor — a kind of protein that attaches itself to DNA — called PcaV was involved in switching the genes on and off. The next step, and the focus of this new research, was figuring out how PcaV controls gene expression.
Sello and Davis proposed a model for how it might work. They proposed that PcaV probably binds to DNA in a way that physically prevents the transcription of the lignin-degrading genes, turning them off. In order to explain how the genes are switched on in the presence of protocatechuate, they proposed that the compound might compromise the ability of PcaV to bind to DNA, which would expose the genes and allow them to be expressed.
A series of experiments provided support for their model. In a test tube, the researchers established that PcaV tightly binds to specific DNA sequences in close proximity to the gene cluster in question, validating the first half of the model. To confirm the second half, Sello and his colleagues exposed the PcaV-DNA complex to protocatechuate. They found that PcaV loses its affinity for DNA in the presence of the compound.
“So we can say that protocatechuate attenuates the DNA binding activity of the PcaV protein, which permits expression of the genes,” Sello said. “We now have evidence that validates our model.”
Sello and his colleagues then dug down into the process a little further. Using a technique called protein crystallography, the team examined the structure of PcaV in complex with protocatechuate, to gain insight into how the compound affects the capacity of the protein to bind DNA. They found that arginine-15, an amino acid in PcaV, is critical for binding both to protocatechuate and to DNA. On this basis, the group proposed that arginine-15 acts as a molecular switch that is flipped by protocatechuate.
“In the absence of protocatechuate, arginine 15 is important in the binding of PcaV to DNA in a manner that represses transcription,” Sello said. “But when you introduce protocatechuate, the arginine spatially reorients to bind the compound. The accompanying conformational changes compromise the ability of the protein to bind to DNA.”
An important protein
Beyond possible implications for bioenergy and sustainable chemistry, Sello says the work is meaningful for basic research in bacteriology. PcaV is a member of the MarR family of transcription factors, a family of 12,000 proteins that are known to regulate genes that influence virulence and drug resistance in other types of bacteria.
“It’s a very important family of transcription factors in bacteria,” Sello said. “Our paper is particularly important because it sheds light on how these proteins control gene expression and mediate responses to small molecules. While many studies of MarR family transcription factors have been published, ours is unique in the way that it synergistically integrates experimental methods from genetics, biochemistry, biophysics, and structural biology.”
The research was funded by the National Science Foundation, the National Institutes of Health, and through a seed award from the Office of the Vice President of Research at Brown.
“The collegiality and size of the scientific community at Brown helps to make these collaborative, interdisciplinary research projects work,” Sello said.
Editors: Brown University has a fiber link television studio available for domestic and international live and taped interviews, and maintains an ISDN line for radio interviews. For more information, call (401) 863-2476.
Media Contact
More Information:
http://www.brown.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Red light therapy for repairing spinal cord injury passes milestone
Patients with spinal cord injury (SCI) could benefit from a future treatment to repair nerve connections using red and near-infrared light. The method, invented by scientists at the University of…
Insect research is revolutionized by technology
New technologies can revolutionise insect research and environmental monitoring. By using DNA, images, sounds and flight patterns analysed by AI, it’s possible to gain new insights into the world of…

X-ray satellite XMM-newton sees ‘space clover’ in a new light
Astronomers have discovered enormous circular radio features of unknown origin around some galaxies. Now, new observations of one dubbed the Cloverleaf suggest it was created by clashing groups of galaxies….