
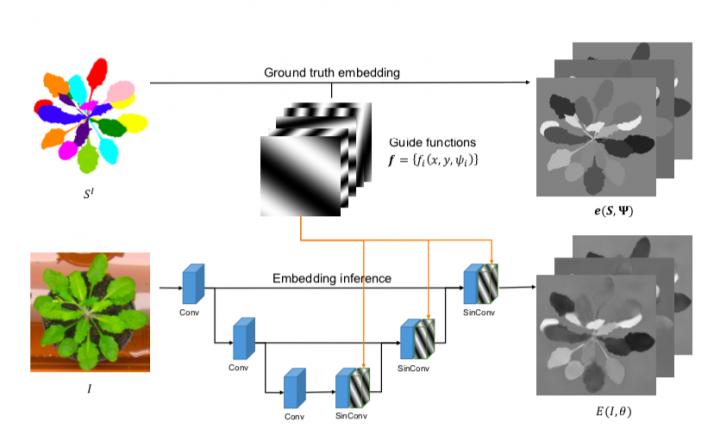
At train-time, we embed each pixel of the ground truth image SI as the mean of predefined guide functions f over instance pixels it belongs to, resuling in embeddings e(S, Ψ). We then train the neural network E to reproduce the ground truth embedding given the input image I. To simplify learning, guide functions f are inputed into intermediate representations of the network using SinConv layers. The learning process uses a simple pixelwise L1-Loss between ground truth embed- ding e(S,Ψ) and the neural network prediction E(I,θ) as a learning objective. At test time instances are retrieved from the predicted embedding E(I, θ) using mean shift clustering.
Credit: Skoltech
Biologists get a wealth of information in the form of biological images, which makes their automatic processing a formidable task.
Researchers often have to handle images with a large number of objects, which is especially hard when it comes to microscopy images with overlapping objects and poor image quality and sharpness.
Machine Learning (ML) helps train the computer to process biological images, making data analysis much faster, more accurate and consistent across experiments.
Skoltech Computer Vision Laboratory of the Skoltech Center for Computational and Data-Intensive Science and Engineering (CDISE) put forward a new method for segmenting biological objects, such as individual cells, organisms and parts of plants, in complex images.
The first author of the study was Victor Kulikov, who worked as a research scientist under the supervision of Skoltech professor Victor Lempitsky. The new method reduces the challenging object separation task to a simpler regression problem.
This is achieved by introducing additional “harmonic” signals into the neural network's input layers and automatically tuning the signals' parameters to the typical size and arrangement of the objects to be isolated.
The scientists used four types of images: photos of plants, pictures with a large number of C. Elegants worms, microscopic images of E. Coli bacteria, and HeLa cancer cell culture images.
Their two-step neural network training algorithm coped brilliantly with the task: trained for a selected image type, the neural networks outperformed other methods in isolating plant leaves, worms, cancer cells, and individual bacteria.
The new method can be employed in scientific research and healthcare applications.
“A major advantage of the new method is that it can learn even from small datasets. We hope that our algorithm will find use in both biological research and other fields where labeled training images are hard to obtain,” Lempitsky said.












