Atomic-scale view of bacterial proteins offers path to new tuberculosis drugs
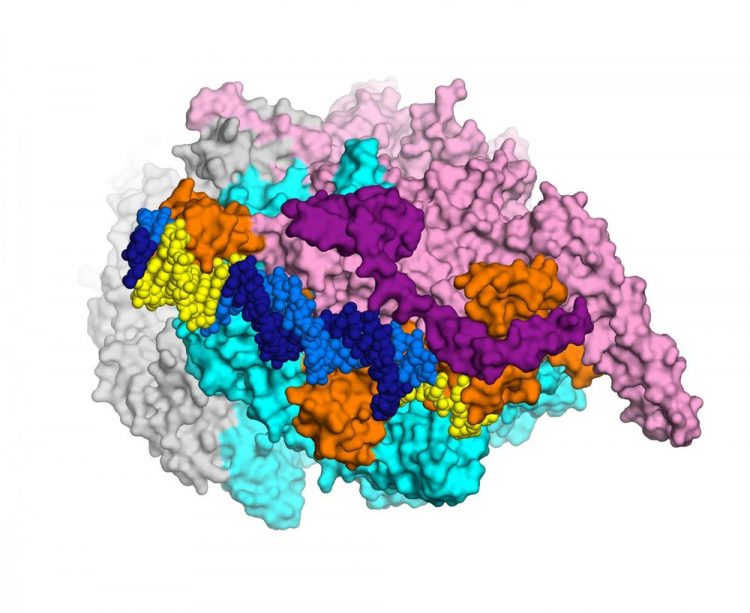
This detailed, 3-D map of a cluster of bacterial proteins is yielding new ideas for how to combat multi-resistant tuberculosis. Credit: Laboratory of Molecular Biophysics at the Rockefeller University/eLife
The research, conducted by a Rockefeller team led by Elizabeth Campbell, in collaboration with scientists at Memorial Sloan Kettering, focuses on a cluster of interacting proteins called RNA polymerase. Crucial to all cells, this protein machine carries out a fundamental process in which genes within the DNA blueprint are copied into RNA. The RNA polymerase is the target of the antibiotic rifampicin–a lynchpin of modern TB treatment, which relies on a combination of drugs. Some bacteria become resistant to rifampicin by acquiring RNA polymerase mutations.
“Now that we can visualize the molecular machinery of the bacteria that the drug targets, we can use a structure-guided approach to better understand how the drug works, how bacteria become resistant to it, and how to potentially improve the drug's action,” says Campbell, a senior research associate in Seth Darst's Laboratory of Molecular Biophysics. She is one of the senior authors of a report published in the online journal eLife.
A molecular map
To visualize the structure the researchers used an imaging method known as x-ray crystallography. By crystalizing enzymes and other molecules interacting with each other–essentially freezing them in action–investigators are able to see how they fit together, much like keys fitting into locks. This ability to visualize what's going on can point the way toward more effective drugs, which may be able to latch more securely onto enzymes and other molecules.
“Based on the findings reported in this study,” Campbell says, “we're already investigating new compounds with new mechanisms of action that appear to inhibit the rifampicin-resistant version of TB. Our eventual goal is to get them into clinical trials investigating new treatments for TB, including rifampicin-resistant TB.”
Rockefeller chemist Sean Brady, who was not directly involved in the study, provided the team with these new compounds. He is now working together with Campbell, Darst, and other colleagues to further develop them into antibiotics and characterize the basis of their activity.
Not all bacteria are alike
It's estimated that up to one-third of the world's population is infected with M. tuberculosis, the bacterium that causes TB. In the study, the researchers worked with a closely related strain called M. smegmatis. “We needed hundreds of liters of cells to get enough of the material to do the crystallization,” says Elizabeth Hubin, a former Rockefeller graduate student who carried out much of the work. “M. tuberculosis grows too slowly to be able to collect the volume that's needed, and it's very dangerous to work with in the lab.”
But M. smegmatis relies on an RNA polymerase that is almost identical in sequence, structure, and behavior to the M. tuberculosis RNA polymerase, which led to another important finding in the study: The RNA polymerase from Escherichia coli, the bacterium most commonly used in lab research, is not. This means there may be a drawback to relying on E. coli as a model when developing certain types of antibiotics for bacteria that cause TB or other diseases.
“Most of the studies previously done with RNA polymerase were done using E. coli,” Campbell says. “We've always assumed that the enzyme works the same way in all bacteria, but our study shows we can't assume what's found in one bacteria applies to all bacteria.
“Every pathogen needs to be studied individually,” she adds, “so the field has a lot of work to do.”
Media Contact
All latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

Silicon Carbide Innovation Alliance to drive industrial-scale semiconductor work
Known for its ability to withstand extreme environments and high voltages, silicon carbide (SiC) is a semiconducting material made up of silicon and carbon atoms arranged into crystals that is…

New SPECT/CT technique shows impressive biomarker identification
…offers increased access for prostate cancer patients. A novel SPECT/CT acquisition method can accurately detect radiopharmaceutical biodistribution in a convenient manner for prostate cancer patients, opening the door for more…

How 3D printers can give robots a soft touch
Soft skin coverings and touch sensors have emerged as a promising feature for robots that are both safer and more intuitive for human interaction, but they are expensive and difficult…