Yeast protein network could provide insights into human obesity
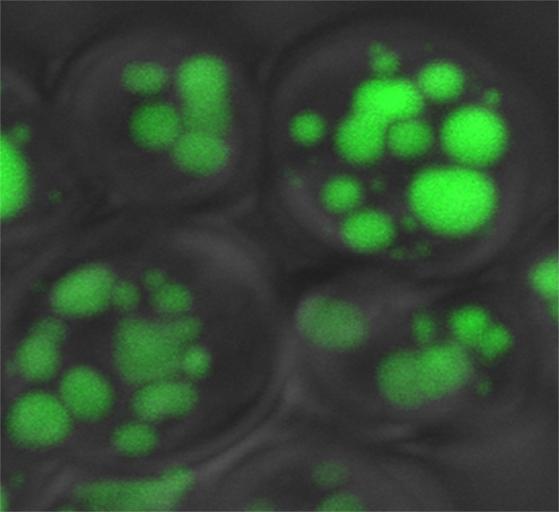
The "fatter" yeast cells that have had SNF4 knocked out. The larger fat droplets are colored green. Courtesy of Patrick Arpp/Caltech
“Removal of any one of the proteins results in an increase in cellular fat content, which is analogous to obesity,” says study coauthor Bader Al-Anzi, a research scientist at Caltech.
The findings, detailed in the May issue of the journal PLOS Computational Biology, suggest that yeast could serve as a valuable test organism for studying human obesity.
“Many of the proteins we identified have mammalian counterparts, but detailed examinations of their role in humans has been challenging,” says Al-Anzi. “The obesity research field would benefit greatly if a single-cell model organism such as yeast could be used–one that can be analyzed using easy, fast, and affordable methods.”
Using genetic tools, Al-Anzi and his research assistant Patrick Arpp screened a collection of about 5,000 different mutant yeast strains and identified 94 genes that, when removed, produced yeast with increases in fat content, as measured by quantitating fat bands on thin-layer chromatography plates. Other studies have shown that such “obese” yeast cells grow more slowly than normal, an indication that in yeast as in humans, too much fat accumulation is not a good thing. “A yeast cell that uses most of its energy to synthesize fat that is not needed does so at the expense of other critical functions, and that ultimately slows down its growth and reproduction,” Al-Anzi says.
When the team looked at the protein products of the genes, they discovered that those proteins are physically bonded to one another to form an extensive, highly clustered network within the cell.
Such a configuration cannot be generated through a random process, say study coauthors Sherif Gerges, a bioinformatician at Princeton University, and Noah Olsman, a graduate student in Caltech's Division of Engineering and Applied Science, who independently evaluated the details of the network. Both concluded that the network must have formed as the result of evolutionary selection.
In human-scale networks, such as the Internet, power grids, and social networks, the most influential or critical nodes are often, but not always, those that are the most highly connected.
The team wondered whether the fat-storage network exhibits this feature, and, if not, whether some other characteristics of the nodes would determine which ones were most critical. Then, they could ask if removing the genes that encode the most critical nodes would have the largest effect on fat content.
To examine this hypothesis further, Al-Anzi sought out the help of a mathematician familiar with graph theory, the branch of mathematics that considers the structure of nodes connected by edges, or pathways. “When I realized I needed help, I closed my laptop and went across campus to the mathematics department at Caltech,” Al-Anzi recalls. “I walked into the only office door that was open at the time, and introduced myself.”
The mathematician that Al-Anzi found that day was Christopher Ormerod, a Taussky-Todd Instructor in Mathematics at Caltech. Al-Anzi's data piqued Ormerod's curiosity. “I was especially struck by the fact that connections between the proteins in the network didn't appear to be random,” says Ormerod, who is also a coauthor on the study. “I suspected there was something mathematically interesting happening in this network.”
With the help of Ormerod, the team created a computer model that suggested the yeast fat network exhibits what is known as the small-world property. This is akin to a social network that contains many different local clusters of people who are linked to each other by mutual acquaintances, so that any person within the cluster can be reached via another person through a small number of steps.
This pattern is also seen in a well-known network model in graph theory, called the Watts-Strogatz model. The model was originally devised to explain the clustering phenomenon often observed in real networks, but had not previously been applied to cellular networks.
Ormerod suggested that graph theory might be used to make predictions that could be experimentally proven. For example, graph theory says that the most important nodes in the network are not necessarily the ones with the most connections, but rather those that have the most high-quality connections. In particular, nodes having many distant or circuitous connections are less important than those with more direct connections to other nodes, and, especially, direct connections to other important nodes. In mathematical jargon, these important nodes are said to have a high “centrality score.”
“In network analysis, the centrality of a node serves as an indicator of its importance to the overall network,” Ormerod says.
“Our work predicts that changing the proteins with the highest centrality scores will have a bigger effect on network output than average,” he adds. And indeed, the researchers found that the removal of proteins with the highest predicted centrality scores produced yeast cells with a larger fat band than in yeast whose less-important proteins had been removed.
The use of centrality scores to gauge the relative importance of a protein in a cellular network is a marked departure from how proteins traditionally have been viewed and studied–that is, as lone players, whose characteristics are individually assessed. “It was a very local view of how cells functioned,” Al-Anzi says. “Now we're realizing that the majority of proteins are parts of signaling networks that perform specific tasks within the cell.”
Moving forward, the researchers think their technique could be applicable to protein networks that control other cellular functions–such as abnormal cell division, which can lead to cancer.
“These kinds of methods might allow researchers to determine which proteins are most important to study in order to understand diseases that arise when these functions are disrupted,” says Kai Zinn, a professor of biology at Caltech and the study's senior author. “For example, defects in the control of cell growth and division can lead to cancer, and one might be able to use centrality scores to identify key proteins that regulate these processes. These might be proteins that had been overlooked in the past, and they could represent new targets for drug development.”
###
The paper is entitled “Experimental and Computational Analysis of a Large Protein Network That Controls Fat Storage Reveals the Design Principles of a Signaling Network.”
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Bringing bio-inspired robots to life
Nebraska researcher Eric Markvicka gets NSF CAREER Award to pursue manufacture of novel materials for soft robotics and stretchable electronics. Engineers are increasingly eager to develop robots that mimic the…

Bella moths use poison to attract mates
Scientists are closer to finding out how. Pyrrolizidine alkaloids are as bitter and toxic as they are hard to pronounce. They’re produced by several different types of plants and are…

AI tool creates ‘synthetic’ images of cells
…for enhanced microscopy analysis. Observing individual cells through microscopes can reveal a range of important cell biological phenomena that frequently play a role in human diseases, but the process of…





















