
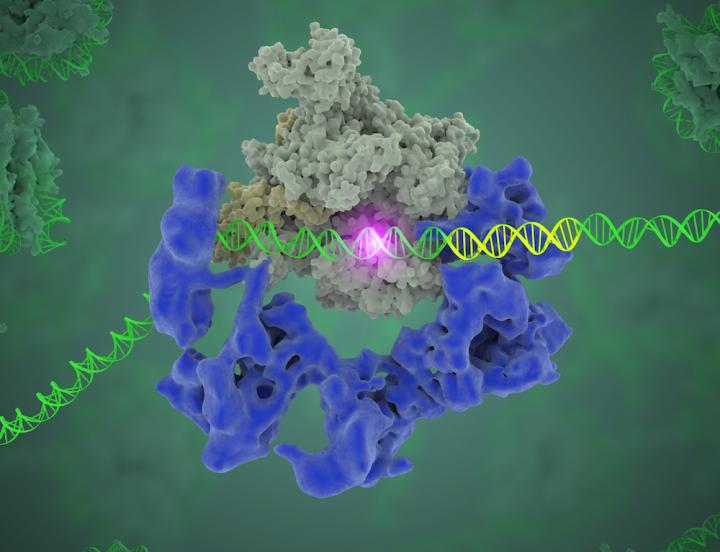
This image shows TFIID (blue) as it contacts the DNA and recruits the polymerase (grey) for gene transcription. The start of the gene is shown with a flash of light.
Credit: Eva Nogales/Berkeley Lab
Your DNA governs more than just what color your eyes are and whether you can curl your tongue. Your genes contain instructions for making all your proteins, which your cells constantly need to keep you alive. But some key aspects of how that process works at the molecular level have been a bit of a mystery–until now.
Using cryo-electron microscopy (cryo-EM), Lawrence Berkeley National Laboratory (Berkeley Lab) scientist Eva Nogales and her team have made a significant breakthrough in our understanding of how our molecular machinery finds the right DNA to copy, showing with unprecedented detail the role of a powerhouse transcription factor known as TFIID.
This finding is important as it paves the way for scientists to understand and treat a host of malignancies. “Understanding this regulatory process in the cell is the only way to manipulate it or fix it when it goes bad,” said Nogales. “Gene expression is at the heart of many essential biological processes, from embryonic development to cancer. One day we'll be able to manipulate these fundamental mechanisms, either to correct for expression of genes that should or should not be present or to take care of malignant states where the process has gone out of control.”
Their study has been published in the journal Nature in an article titled, “Structure of promoter-bound TFIID and insight into human PIC assembly.” The lead author is Robert Louder, a biophysics graduate student in Nogales' lab, and other authors are Yuan He, José Ramón López-Blanco, Jie Fang, and Pablo Chacón.
Nogales, a biophysicist who also has appointments at Howard Hughes Medical Institute and UC Berkeley, has been studying gene expression for 18 years. While she and her team have made several significant findings in recent years, she calls this the biggest breakthrough so far. “This is something that will go in biochemistry textbooks,” she said. “We now have the structure of the whole protein organization that is formed at the beginning of every gene. This is something no one has come close to doing because it is really very difficult to study by traditional methodologies.”
How genetic information flows in living organisms is referred to as the “central dogma of molecular biology.” Cells are constantly turning genes on and off in response to what's happening in their environment, and to do that, the cell uses its DNA, the big library of genetic blueprints, finds the correct section, and makes a copy in the form of messenger RNA; the mRNA is then used to produce the needed protein.
The problem with this “library” is that it has no page numbers or table of contents. However, markers are present in the form of specific DNA sequences (called core promoter motifs) to indicate where a gene starts and ends. So how does the polymerase, the enzyme that carries out the transcription, know where to start? “DNA is a huge, huge molecule. Out of this soup, you have to find where this gene starts, so the polymerase knows where to start copying,” Nogales said. “This transcription factor, TFIID, is the protein complex that does exactly that, by recognizing and binding to DNA core promoter regions.”
What Nogales and her team have been able to do is to visualize, with unprecedented detail, TFIID bound to DNA as it recognizes the start, or promoter, region of a gene. They have also found how it serves as a sort of landing pad for all the molecular machinery that needs to assemble at this position–this is called the transcription pre-initiation complex (PIC). This PIC ultimately positions the polymerase so it can start transcribing.
“TFIID has to do not only the binding of the DNA, recruitment, and serving as landing pad, it has to somehow do all that differently for different genes at any given point in the life of the organism,” Nogales said.
Added Louder: “We have generated the first ever structural model of the full human TFIID-based PIC. Our model yields novel insights into human PIC assembly, including the role of TFIID in recruiting other components of the PIC to the promoter DNA and how the long observed conformational flexibility of TFIID plays a role in the regulation of transcription initiation.”
Proteins have traditionally been studied using X-ray crystallography, but that technique has not been possible for this kind of research. “TFIID has not been accessible to protein crystallography because there's not enough material to crystallize it, it has very flexible elements, and it is of a huge size,” Nogales said. “All of those things we can overcome through cryo-EM.”
Cryo-EM, in which samples are imaged at cryogenic temperatures without need for dyes or fixatives, has been used since the 1980s in structural biology. With extensive computational analysis of the images researchers are able to obtain three-dimensional structures. However, cryo-EM has undergone a revolution in the last few years with the advent of new detectors–developed, in fact, at Berkeley Lab–that improve resolution and reduce the amount of data needed by up to a hundred-fold.
“Many biological systems we had thought were impossible to study at high resolution have become accessible,” she said. “Now the resolution allows us to get atomic details. This is an area in which Berkeley Lab has been one of the leaders.”
While this study has revealed important new insights into gene expression, Nogales notes that much work remains to be done. Next she plans to investigate how TFIID is able to recognize different sequences for different gene types and also how it is regulated by cofactors and activators.
“We are just at the beginning,” she said. “This complex, TFIID, is very, very critical. Now we have broken barriers in the sense that we can start generating atomic models and get into details of how DNA is being bound.”
###
This research was supported by the National Institutes of Health's National Institute of General Medical Sciences and by the Spanish Ministry of Economy and Competitiveness. Computational work was carried out at the National Energy Research Scientific Computing Center (NERSC), a DOE Office of Science User Facility hosted at Berkeley Lab. Nogales is a Senior Faculty Scientist in Berkeley Lab's Molecular Biophysics and Integrated Bioimaging Division.
Lawrence Berkeley National Laboratory addresses the world's most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab's scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy's Office of Science. For more, visit http://www.
DOE's Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.












