UMD-led study identifies the off switch for biofilm formation
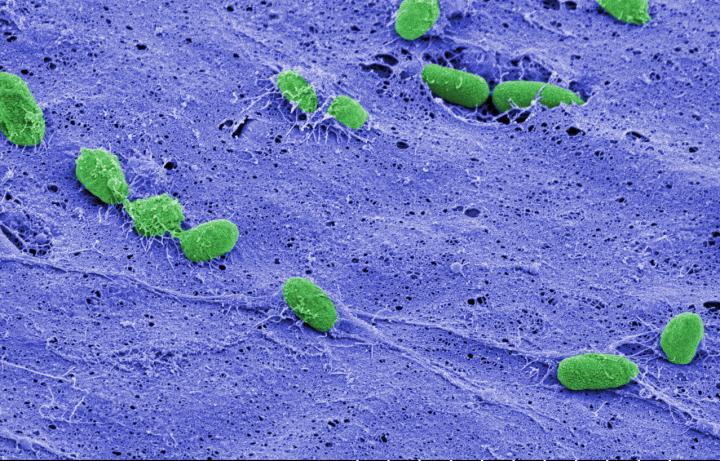
In this false-colored image, individual cells of Pseudomonas aeruginosa (green) can be seen resting on the fibrous surface of a biofilm (purple) that helps protect cells beneath its surface. At top right, two cells incorporated within the biofilm peek out from a fissure in the film's surface. Credit: Debra Weinstein, Sao-Mai Nguyen-Mau, and Vincent Lee
Bacteria are best known as free-living single cells, but in reality their lives are much more complex. To survive in harsh environments, many species of bacteria will band together and form a biofilm–a collection of cells held together by a tough web of fibers that offers protection from all manner of threats, including antibiotics. A familiar biofilm is the dental plaque that forms on teeth between brushings, but biofilms can form almost anywhere given the right conditions.
Biofilms are a huge problem in the health care industry. When disease-causing bacteria establish a biofilm on sensitive equipment, it can be impossible to sterilize the devices, raising rates of infection and necessitating expensive replacements. So researchers look for ways to break down the defenses of biofilms to prevent them from establishing a foothold.
Now, a University of Maryland-led team has found an important link in the biofilm formation process: an enzyme that shuts down the signals that bacteria use to form a biofilm. The findings, reported in the August 24, 2015 Early Online Edition of the Proceedings of the National Academy of Sciences, have far-reaching implications for the development of new treatments, and could one day help make biofilm-related complications a distant memory.
“Bacteria form biofilms because they sense a change in their environment. They do this by generating a signaling molecule, which binds to a receptor that turns on the response,” said Mona Orr, the lead author of the study and a UMD biological sciences graduate student. “But you need a way to turn off the switch–to remove the signal when it's no longer needed. We've identified the enzyme that completes the process of turning off the switch.”
The well-known switch that activates biofilm formation is a signaling molecule called Cyclic-di-GMP, also known as c-di-GMP. Many species of disease-causing bacteria use c-di-GMP to signal the formation of biofilms, including Escherichia coli, Salmonella enterica and Vibrio cholerae.
But Orr and her colleagues are the first to identify the molecule that completes the process of clearing c-di-GMP from the cell, thus ending the biofilm signaling process. The molecule is an enzyme called oligoribonuclease, and much like c-di-GMP, oligoribonuclease is also common among disease-causing bacterial species.
The team studied the process in the bacteria Pseudomonas aeruginosa, a common species known to cause infections in hospital patients. But because of the genetic and physiological similarities between P. aeruginosa and other infectious species, the researchers believe that oligoribonuclease serves the same function across a wide variety of bacteria.
“You can think of this process in terms of water filling a sink. The rate of water from the faucet is just as important as the size of the drain in determining the level of water in the sink,” said Vincent Lee, a co-author of the study and an associate professor in the UMD Department of Cell Biology and Molecular Genetics and the Maryland Pathogen Research Institute. “The level of c-di-GMP in the cell is analogous to the amount of water in the sink. Because no one knew what the drain was, our findings create a complete picture of the signaling process.”
Orr, Lee and their colleagues from the UMD Department of Chemistry and Biochemistry and Michigan State University focused their work on P. aeruginosa because it is well studied and can survive under a variety of conditions, making it notoriously difficult to control. Contact lens wearers might already be familiar with P. aeruginosa, as it commonly forms infectious, green-tinted biofilms on older lenses or those that have not been cleaned properly.
The team found that oligoribonuclease is necessary for the second of a two-step process. The first, which converts c-di-GMP into an intermediate molecule called pGpG, was already known. Orr, Lee and their colleagues have now filled in the important second step in this process: oligoribonuclease breaks apart pGpG and thus completely shuts off the signaling pathway.
The result suggests that oligoribonuclease could be used to help design new antibiotics, disinfectants, and surface treatments to control biofilms. Such measures could prevent infections and preclude the need for frequent replacement of expensive hospital equipment. Because biofilms can also form on implanted medical devices, such as pacemakers and synthetic joints, effective treatments against biofilms could eliminate the need for costly and risky replacement surgeries.
While oligoribonuclease most likely shuts down biofilm formation in many infectious bacterial species, Orr and Lee acknowledge that their discovery is not quite a “silver bullet” that can fight every type of biofilm.
“The genes that make these signals are found in most bacteria. The oligoribonuclease enzyme that breaks the effect is only found in some, however,” Lee explained. “So there must be parallels in the organisms that don't have oligoribonuclease. Finding these other 'off' switches is high on our list of future research goals.”
###
In addition to Orr and Lee, UMD authors on the paper include Herman Sintim, associate professor in the UMD Department of Chemistry and Biochemistry, Jingxin Wang (Ph.D. '11, chemistry) and Gregory Donaldson (B.S. '10, biological sciences).
This work was funded by the National Institutes of Health's National Institute of Allergy and Infectious Diseases (Award Nos. T32-AI089621 and R21AI096083) and the National Science Foundation (Award Nos. MCB1253684 and CHE0746446). The content of this article does not necessarily reflect the views of these organizations.
The research paper, “Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover,” Mona Orr, Gregory Donaldson, Geoffrey Severin, Jingxin Wang, Herman Sintim, Christopher Waters, and Vincent Lee, was published August 24, 2015 in the Early Online Edition of the Proceedings of the National Academy of Sciences.
Media Relations Contact: Matthew Wright, 301-405-9267, mewright@umd.edu
University of Maryland
College of Computer, Mathematical, and Natural Sciences
2300 Symons Hall
College Park, MD 20742
http://www.
@UMDscience
About the College of Computer, Mathematical, and Natural Sciences
The College of Computer, Mathematical, and Natural Sciences at the University of Maryland educates more than 7,000 future scientific leaders in its undergraduate and graduate programs each year. The college's 10 departments and more than a dozen interdisciplinary research centers foster scientific discovery with annual sponsored research funding exceeding $150 million.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















