
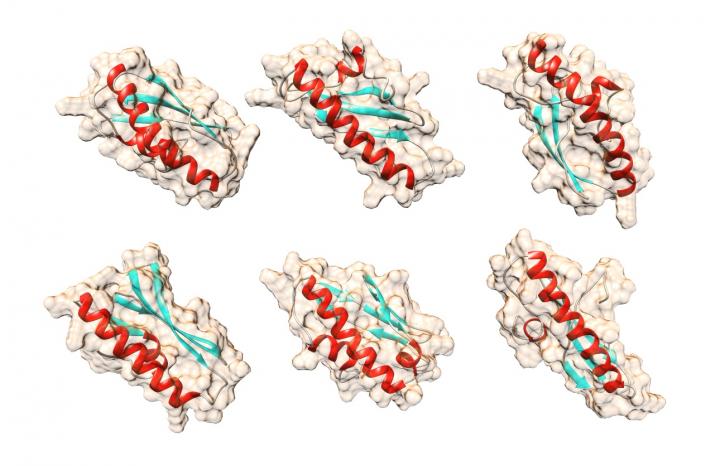
Possible shapes for the yeast protein Bsc4 that the computer algorithm QUARK predicted using only the protein's amino acid sequences.
Credit: Matthew Cordes, using UCSF Chimera molecular graphics package
Usage Restrictions: The image may only be used to illustrate a story about the research described in the accompanying news release, "The Birth of a New Protein." Please make sure to credit the image as stated. Do not post this image independent of the story.
Scientists thought such newly evolved proteins were works-in-progress that could not fold into complex shapes the way more ancient proteins do.
“This new study from our laboratory provides the very first evidence that a protein evolved recently from scratch can in fact fold into a compact three-dimensional shape,” said senior author Matthew Cordes, a UA associate professor of chemistry and biochemistry.
“What we've discovered doesn't come across as a completely developed biological molecule. It looks like what you'd think a newborn protein would look like — maybe a little clumsy or hazily formed,” said Cordes, a member of the UA's BIO5 Institute. “But nonetheless, this protein folds into a three-dimensional structure and by a lot of measures it looks like proteins that evolved long ago.
“It's astonishing when I look at it. I really feel like I'm seeing something hatch out.”
Recent evidence suggests new genes can arise from the non-coding sections, or “junk,” DNA and that those new genes could code for brand-new proteins. Genes code for proteins, the molecules that do work inside cells.
Co-author Joanna Masel said, “Not all genes are ancient and the ones that aren't ancient don't get studied a lot.”
Cordes is the first person to look at the structure of the protein that results from a gene that recently arose from junk DNA, said Masel, a UA professor of ecology and evolutionary biology and a member of the BIO5 Institute.
Because changing an existing gene seemed easier than inventing an entirely new one, scientists had thought new genes arose by modifying duplicates of existing genes.
“The story is that evolution is not just a tinkerer that reworks what it has–sometimes evolution invents a completely new functional, structured molecule,” said Cordes.
The team's research paper, “Foldability of a Natural De Novo Evolved Protein,” is online in the journal Structure and scheduled for publication in November. Cordes' and Masel's co-authors are Dixie Bungard, Jacob Copple, Jimmy Chhun, Vlad Kumirov and Scott Foy of the UA and Jing Yan and Vicki Wysocki of Ohio State University in Columbus.
The National Institutes of Health grants GM104040, GM113658, ES06694, CA023074 and 1S10 RR028868-01 and a grant from the John Templeton Foundation funded the research.
Scientists have known for more than a decade that some genes appear to arise brand-new, or de novo, from junk DNA.
However, little is known about the proteins such genes code for, said Cordes, who studies the evolution of protein structure and function.
“My goal is to see what these proteins look like,” he said.
Cordes and his colleagues studied a protein that a laboratory strain of baker's yeast makes using a de novo gene. The gene BSC4 and its resulting protein Bsc4 seem to help in DNA repair.
The first step in studying a protein is making a lot of it. The Cordes team used a standard method: They inserted the BSC4 gene into E. coli bacteria and had the bacteria serve as a protein-making factory. Some pharmaceutical companies use the same method to make insulin for people with diabetes.
The next step is purifying the protein, which for Bsc4 had never been done before, Cordes said.
“There's been a real lack of people purifying the products of these de novo genes and examining them. That's the gap we're trying to fill,” he said.
Ordinarily, researchers then crystallize the protein and make a detailed description of the protein's structure using high-resolution imaging techniques.
However, the baby protein wasn't easy to work with, Cordes said. The team could not crystallize it.
The team did determine general information about the protein's shape and structure. The BSC4 gene codes for one small chain of amino acids, or polypeptide chain.
In the resulting protein, Bsc4, several of those chains come together. The protein can exist as two or three chains, but also as groups of many chains. Having such a variety of sizes is not typical, Cordes said.
“Most natural proteins exist in a certain size,” he said. “This one can be in groups of four, five, six, seven – that's one reason you might call the structure fuzzy or rudimentary.”
The protein does exhibit other characteristics of normal proteins, including folding into standard shapes such as alpha helices and beta-pleated sheets. In addition, the protein will unfold, or denature, under certain conditions and can then be coaxed back into its refolded state. The firm white portion of a cooked egg is denatured egg-white protein.
“For many years, people thought that proteins had to have these elegant origami-like structures to function but it has become clear that they can have disorder, they can have molten properties and still do things in an organism,” he said.
Cordes characterized Bsc4 as “globular” and said most natural globular proteins have just one standard shape. He calls the protein “molten globular” because it may shift between different shapes.
“The protein is thought to be involved in DNA repair and might be involved in binding DNA. It may be that its current structure is perfectly good at what it needs to do,” he said.
“Not having emerged as this glorious work of art makes it more believable that these protein-coding genes are popping up out of the genetic region between genes,” Cordes said. “I want to know where things come from in life – how life creates novelty, how evolution creates novelty.”
###
Researcher contact:
Matthew Cordes
520-626-1175
cordes@email.arizona.edu
Joanna Masel
520-626-9888
masel@email.arizona.edu












