Stalking a Wily Foe: Scientists Figure Out How C. difficile Bacteria Wreak Havoc in Gut
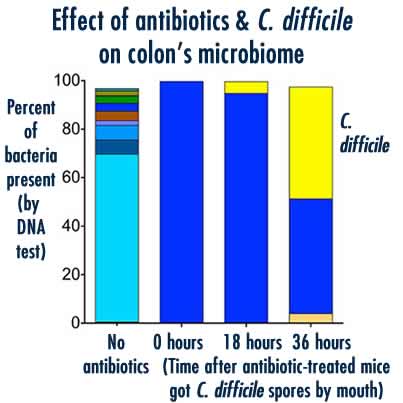
Young lab, University of MIchigan Medical School The combination of antibiotics and C. difficile infection completely changed the microbiome - or community of bacteria - in the colon. The left column shows the DNA of many species of bacteria was present without antibiotics; the right three show how first antibiotics, and then C. difficile, changed the population.
Sometimes, science means staying awake for two days straight.
But losing sleep is a small sacrifice to make, if you want to learn more about tiny bacteria that sicken half a million Americans each year, kill more than 14,000 of them, and rack up $4.8 billion in health care costs.
That’s what drove a team of University of Michigan scientists to work around the clock to study the bacterium called Clostridium difficile, or C. difficile, the bane of hospitals and nursing homes. Most patients develop it after taking antibiotics.
In a new paper in the journal Infection and Immunity, the researchers lay out for the first time exactly how C. difficile wreaks havoc on the guts of animals in a short time, and causes severe diarrhea and life-threatening disease in humans.
Despite the heavy toll the organism takes, no team had ever been able to measure C. difficile activity over time in this way. Their findings could help lead to better prevention and treatment of C. difficile infections.
A fast track to disease
The researchers started by introducing C. difficile spores into mice via their mouths – similar to what might happen in a hospital environment where spores from past patients’ infections abound. Then, they studied what happened after the spores entered the body, by taking gut samples at regular intervals and studying them under special conditions. The animals had all received antibiotics.
Through their hours-long surveillance, the researchers found that it took C. difficile only about 24 hours to go from hard spores to toxin-producing, diarrhea-inducing cells all the way at the other end of the digestive tract, in the large intestine.
The researchers also show that bile acids in the gut “woke up” the dormant bacteria spores, and that they grow into cells in the small intestine within 24 hours of exposure. Because antibiotics disrupted the gut’s normal community of other bacteria – called the gut microbiome — C. difficile cells could continue to the large intestine and start their toxic effects on the cells that line the gut. When they tested the contents of the small intestine separately, they also showed this happens whether or not the animals have received antibiotics.
In the large intestine, they even saw how C. difficile cells formed spores again – allowing them to survive the exit from the body in feces and go on to infect a new host.
“If we can understand the process that specific bacteria use to germinate and get established, we may be able to intervene more effectively,” says Vincent Young, M.D., Ph.D., the senior author of the new study, a professor at the U-M Medical School and co-leader of the school’s Host Microbiome Initiative. “We assume that antibiotics change the gut microbiome, but we haven’t known how that allows C. difficile to gain a foothold and begin to ramp up growth.”
First author Mark Koenigsknecht, Ph.D., a postdoctoral fellow in Young’s lab who is now continuing his research at the U-M College of Pharmacy, was one of the researchers who was up all night to get data for the experiment.
“We introduced 100 spores through the mouth, and within six hours we could find 1,000 cells in the intestinal tract,” he notes. “We chose this strain of C. difficile because of its rapid ability to cause disease in animals, but we didn’t think it would happen that quickly.”
Tracking C. difficile’s effect on the gut
The U-M team used a mouse model they developed, and a common antibiotic in the cephalosporin class. The strain of C. difficile used in the experiment originated with a patient years ago, but is available for purchase as a laboratory culture.
Working in facilities made possible by the Host Microbiome Initiative, they took samples at regular intervals from seven difficileerent areas of the digestive tracts of the mice. They then whisked the samples into special oxygen-free facilities, called anaerobic chambers, that allowed them to see the amount and forms of C. difficile present in each gut region.
With the help of Patrick Schloss, Ph.D., a professor in the Department of Microbiology & Immunology, the researchers used DNA analysis to see what the entire gut microbiome looked like in antibiotic-treated animals and those that hadn’t been treated. The antibiotics really disrupted the community of bacteria in the small intestine, and C. difficile came to dominate in 36 hours.
They also examined the intestinal tract under a microscope. The toxin produced by C. difficile cells in their vegetative, or growing, state causes an effect on the cells that line the digestive tract, causing them to become “leaky”, raising the alarm among nearby immune system cells, and leading to diarrhea. The cell changes were seen in the large intestine about 30 hours after spore introduction.
Next steps
Koenigsknecht notes that this is the first time researchers have seen in a living animal that toxin production, and production of new spores of C. difficile capable of surviving outside the body, occur at the same time. This indication that the two processes are linked, and that they are switched on by some factor in the body, is intriguing, he says.
Now, the effort to figure out what that signal is, whether different strains of C. difficile act differently, and who is most vulnerable to its effects, will continue.
Koenigsknecht has teamed with College of Pharmacy professors to test the use of a seven-foot-long tube that can be threaded down the human digestive tract and used to retrieve samples at different locations along the way. Originally developed for testing how drugs are broken down and used by the body, it could provide an entirely new window into the human microbiome.
“Now that we understand what C. difficile is doing, we can also go and ask more questions about how the machinery inside the cell is turning on,” he says. “We have to know how to study it before we can cure it.” Animal-based research is vital to this effort.
Young notes that there are many ways C. difficile could take over an antibiotic-decimated gut. “Does it prevent other bacteria from growing, or outcompete them by eating faster? Does it communicate with the cells of the gut lining? We’re trying to figure out the interaction between the ‘good bugs’ and the ‘bad bugs’, and the lining of the gut.” Young is an associate professor of infectious diseases and of microbiology and immunology.
In addition to Young, Schloss and Koenigsknecht, the study’s authors are Casey Theriot, Ingrid Bergin and Cassie Schumacher. The research was funded by the National Institutes of Health grants U19AI090871, K01GM109236 and 5R01GM099514. It used the U-M Metabolomics Core, funded by NIH grant U24 DK097153. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reference: Infection and Immunity, doi: 10.1128/IAI.02768-14, March 2015 vol. 83 no. 3 934-941
Contact Information
Kara Gavin
Lead Public Relations Representative
kegavin@med.umich.edu
Phone: 734-764-2220
Mobile: 734-516-5362
Media Contact
More Information:
http://www.umich.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















