
Rice University lab runs crowd-sourced competition to create 'big data' diagnostic tools
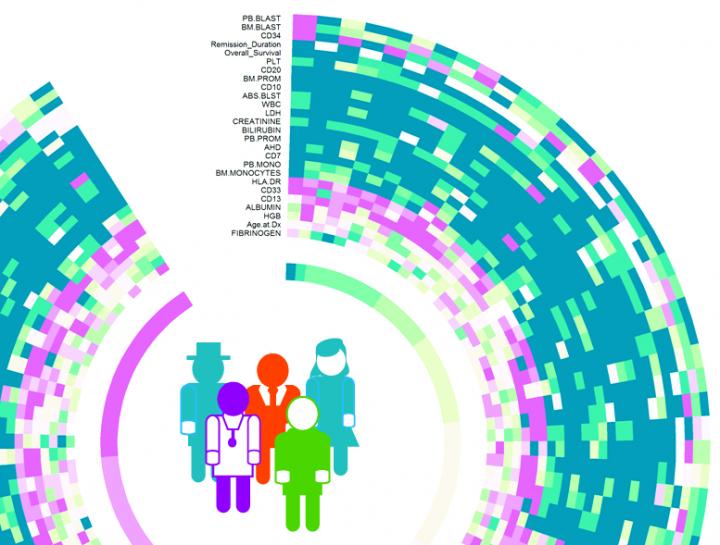
A crowdsourced collaboration/competition known as DREAM 9 that is centered at Rice University set out three years ago to develop ideas for computational tools that would help treat patients with acute myeloid leukemia. The results were announced this week.
Credit: David Noren/Rice University
In the DREAM 9 challenge, 31 teams of computational researchers applied competing methods to a unique set of patient data gathered from hundreds of patients with acute myeloid leukemia at the University of Texas MD Anderson Cancer Center.
Rice bioengineer Amina Qutub is principal investigator of the open-source paper published today in PLOS Computational Biology. Rice served as the competition hub, in line with the university's strategic initiative to foster bioscience collaborations with fellow Texas Medical Center institutions.
DREAM, which stands for Dialogue for Reverse Engineering Assessment and Methods, is a platform for crowd-sourced studies that focus on developing computational tools to solve biomedical problems. Essentially, it's a competition that serves as a large, long-standing, international scientific collaboration.
Acute myeloid leukemia presented a worthy challenge since there is no single genetic cause of the disease, which makes it hard to select treatments for patients suffering from the deadly cancer of the blood, Qutub said.
The DREAM 9 patient data set was collected by Steven Kornblau, a leukemia doctor and professor at MD Anderson. The data was distributed to DREAM 9 participants online through Sage Bionetworks' Synapse web portal and through Biowheel, a cloud-based technology launched by the Qutub Lab.
Biowheel is an interactive tool to visualize and group high-dimensional data of all kinds. It was developed by Rice graduate student Chenyue Wendy Hu, undergraduate alumnus Alex Bisberg and Qutub. National Library of Medicine postdoctoral fellow David Noren and research scientist Byron Long, also of the Qutub Lab, are lead authors of the paper.
For DREAM 9, each team was presented with training data from 191 patients that included demographic information like age and gender and more complex proteomic and phosphoprotein data that describes signaling protein pathways believed to play a role in the disease.
The competition used a test data set from 100 patients that didn't include outcomes, such as whether patients responded to therapy, relapsed, survived or died.
The primary challenge was to see how well the teams' algorithms could predict how patients responded to chemotherapy. The eventual goal is to give clinicians a predictive tool to develop individualized treatment plans.
The top-performing models were by Team EvoMed (Li Liu) of Arizona State University and Team Chipmunks (Honglei Xie, Greg Chen, Xihui Lin, Geoffrey Hunter) of the Ontario Institute for Cancer Research, Toronto. They were best able to predict patient response to therapy with an accuracy of close to 80 percent, Qutub said.
She noted that one interesting takeaway was that overall the 31 models found it harder to predict outcomes for patients classified as “resistant to therapy” than for responsive patients. The median model prediction accuracy for resistant patients was 42 percent vs 73 percent for responsive patients. The winning models were impacted by the perturbation of signaling proteins known as phosphoinositide-3-kinase (a cell-cycle regulator) and NPM1 (which contributes to ribosome assembly and chromatin regulation), singling them out as strong candidates for further study.
The Qutub Lab became involved in leading DREAM 9 after the design of Biowheel won a DREAM 8 subchallenge three years ago. Five Qutub Lab members contributed predictive algorithms to the earlier challenge, which focused on proteomic analysis of breast cancer from MD Anderson data.
In discussions with DREAM organizer Gustavo Stolovitzky of IBM, Qutub had suggested a challenge based on one of the leukemia data sets Kornblau and her lab were analyzing to help understand molecular signaling in cancer.
“We used DREAM as a way to get general insight into making more accurate predictive models of clinical outcomes,” Qutub said. “Steve (Kornblau), who runs the core banking facility for leukemia patients at MD Anderson Cancer Center, had the foresight to start gathering and banking patient biopsy samples when he was a resident over 25 years ago. The bank is a fantastic resource and a tremendous gift to the public. Genomic and proteomic analysis on a portion of these patient biopsies served as the basis for DREAM.”
Because judging the entries was so computationally demanding, the Qutub Lab enlisted Erik Engquist, a co-author of the paper and director of the Center for Research Computing, and Rice's Ken Kennedy Institute for Information Technology (K2I) to help direct data traffic. Engquist helped the lab ensure a level playing field as competitors' algorithms ran on several of the university's high-performance computing platforms. He also helped set up a server to share challenge data via Biowheel, Qutub said.
“We had more than 270 participants and several dozen models to vet. K2I was instrumental in helping us run the challenge,” she said.
Before DREAM 9 began, Noren spent considerable time designing the challenge and processing the complex patient data set. During and following the challenge, Noren, Long and the IBM team spent months processing the mountain of output data so the models, which analyzed 40 clinical indicators and 231 gene-expression profiles for each patient, would get a fair comparison. (The Rice lab did not compete because, as administrator, it already knew the results.)
Noren's task was to compare how well each model performed for each patient and to see whether the top-performing models had unique input parameters or features, Qutub said. “This way, we can start to learn which features of patients uniquely predict their outcomes.”
The results still only hinted at the complexity of determining an optimal leukemia treatment plan, she said. Qutub's lab is using what it learned from its DREAM experience as a basis for experimentation on leukemia cell lines and test whether targeting specific sets of proteins offer a therapeutic advantage.
###
Co-authors of the paper include Rice graduate student André Schultz, also of the Qutub Lab; Raquel Norel and Kahn Rrhissorrakrai of the IBM Computational Biology Center, Yorktown Height, N.Y.; Kenneth Hess of MD Anderson; Paul Boutros of the Ontario Institute for Cancer Research and the University of Toronto, Canada; Oleg Stepanov of the Institute of Systems Biology, Moscow; Thea Norman and Stephen Friend of Sage Bionetworks; and members of the DREAM 9 AML Consortium, along with Noren, Long, Hu, Bisberg, Engquist and Qutub of Rice; Kornblau of MD Anderson; Stolovitzky of IBM and the Icahn School of Medicine at Mount Sinai, N.Y., and members of the winning teams.
Read the paper at http://journals.
This news release can be found online at http://news.
Follow Rice News and Media Relations via Twitter @RiceUNews
Related materials:
DREAM 9 outline: https:/
Qutub Lab: https:/
Rice Department of Bioengineering: http://bioe.
DiBS (Biowheel): http://www.
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation's top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,910 undergraduates and 2,809 graduate students, Rice's undergraduate student-to-faculty ratio is 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for best quality of life and for lots of race/class interaction by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger's Personal Finance. To read “What they're saying about Rice,” go to http://tinyurl.












