
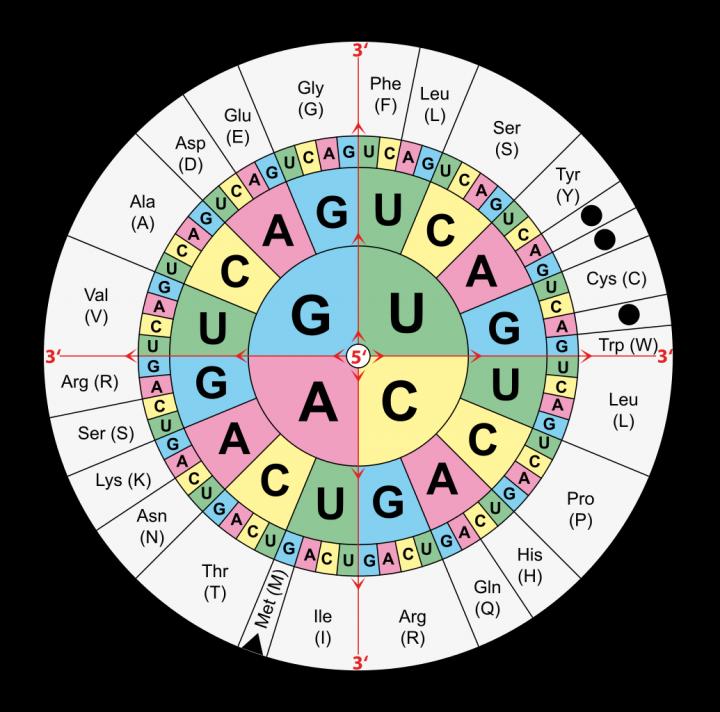
Starting from the four innermost letters and working to the outermost ring, this table shows shows which three-letter base sequence or codon encodes which amino acid. In the journal Angewandte Chemie International Ed., researchers from the US Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, and Yale University have discovered that microorganisms recognize more than one codon for the rare, genetically encoded amino acid selenocysteine.
Credit: Wikimedia Commons
Four letters – A, C, G and T – stand in for the four chemical bases that store information in DNA. A sequence of these same four letters, repeating in a particular order, genetically defines an organism. Within the genome sequence are shorter, three-letter codons that represent one of the 20 regularly used amino acids, with three of the possible 64 three-letter codons reserved for stop signals.
These amino acids are the building blocks of proteins that carry out a myriad of functions. For example, the amino acid alanine can be represented by the three-letter codon GCU and the amino acid cysteine by the three-letter codon UGU. In some organisms, the three-letter codon UGA, which normally signals the end of a protein-coding gene, is hijacked to code for a rare genetically encoded amino acid called selenocysteine.
Published ahead online March 16, 2016 in the journal Angewandte Chemie International Ed., researchers from the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, and Yale University have discovered that microorganisms recognize more than one codon for selenocysteine. The finding adds credence to recent studies indicating that an organism's genetic vocabulary is not as constrained as had been long held.
The work is a follow-up to two 2014 publications; a Science paper by the JGI group finding that some organisms interpret the three “stop” codons which terminate translation to mean anything but. A synthetic biology experiment of the Yale group published in an Angewandte Chemie International Ed. paper revealed the astonishing fact that almost all codons in Escherichia coli could be replaced by selenocysteine. This posed the question whether the same phenomenon can also occur in nature.
“Access to the tremendous resources at the JGI allowed us to quickly test challenging hypotheses generated from my research projects that have been supported over the long-term by DOE Basic Energy Sicences and the National Institutes of Health,” said Dieter Soll, Sterling Professor of Molecular Biophysics and Biochemistry Professor of Chemistry at Yale, the lead author of the paper.
Thus a fruitful collaboration resulted; the combined team scanned trillions of base pairs of public microbial genomes and unassembled metagenome data in the National Center for Biotechnology Information and the DOE JGI's Integrated Microbial Genomes (IMG) data management system to find stop codon reassignments in bacteria and bacteriophages. Delving into genomic data from uncultured microbes afforded researchers the opportunity to learn more about how microbes behave in their natural environments, which in turn provides information on their management of the various biogeochemical cycles that help maintain the Earth.
From approximately 6.4 trillion bases of metagenomic sequence and 25,000 microbial genomes, the team identified several species that recognize the stop codons UAG and UAA, in addition to 10 sense codons, as acceptable variants for the selenocysteine codon UGA.
The findings, the team reported, “opens our minds to the possible existence of other coding schemes… Overall our approach provides new evidence of a limited but unequivocal plasticity of the genetic code whose secrets still lie hidden in the majority of unsequenced organisms.”
This finding also illustrates the context-dependency of the genetic code, that accurately “reading” the code (and interpreting DNA sequences) and ultimately “writing” DNA (synthesizing sequences to carry out defined functions in bioenergy or environmental sciences) will require study of the language of DNA past the introductory course level.
###
This work was enabled by resources from the DOE Joint Genome Institute's Community Science Program (CSP). The CSP annual call for letters of intent are due April 7 and is focused on large-scale sequence-based genomic science projects that address questions of relevance to DOE missions in sustainable biofuel production, global carbon cycling, and biogeochemistry. For more information, see: http://bit.












