
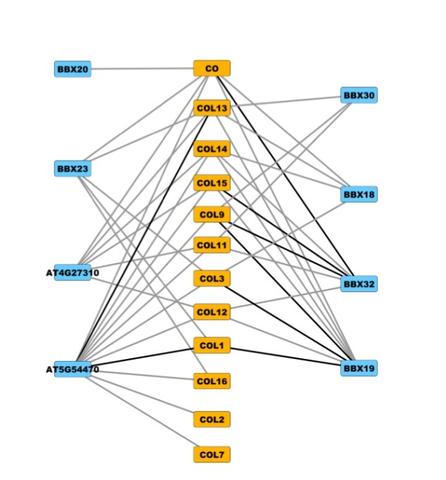
A new mapping method let researchers discover new links (gray lines) between two groups of plant proteins (yellow and blue) that have a common structure (the BBX domain), suggesting many different combinations of interactions, rather than a few, are involved in coordinating cellular programs like flowering time and circadian rhythm.
Creditj: Salk Institute
“The power of this new approach is in the ability we now have to scale it up,” says senior author Joseph Ecker, professor and director of Salk's Genomic Analysis Laboratory and investigator of the Howard Hughes Medical Institute. “This assay has the potential to begin to address questions about fundamental biological interactions that we haven't been able to address before.”
The interactome of a cell, like a map of social networks, lets scientists see who's working with who in the world of proteins. This helps them figure out the roles of different proteins and piece together the different players in molecular pathways and processes.
If a newly discovered protein interacts with lots of other proteins involved in cellular metabolism, for instance, researchers can deduce that's a likely role for the new protein and potentially target it for treatments related to metabolic dysfunction.
Until now, researchers have typically relied on standard high-throughput yeast two-hybrid (Y2H) assays to determine the interactions between proteins. The system requires using a single known protein–known as the “bait”–to screen against a pool of “prey” proteins. But finding all the interactions between, for instance, 1,000 proteins, would require 1000 separate experiments to screen once for each bait's interaction partners.
“Current technologies essentially require that interactions detected in primary screening get retested individually,” says Shelly Trigg, an NSF Graduate Research Fellow at the University of California, San Diego, in the Ecker lab, and first author of the new paper. “That may no longer be necessary with the screening depth this approach achieves.”
In their new method, Ecker, Trigg and their colleagues added a twist to the standard Y2H assay for a much more effective way of measuring the interactome. The genes for two proteins, each on their own circle of DNA, are added to the same cell. If the proteins of interest interact inside the cell, a gene called Cre is activated. When turned on, Cre physically splices the two individual circles of DNA together, thus pairing the genes of interacting proteins together so the team can easily find them through sequencing.
The team can generate a massive library of yeast cells–each containing different pairs of proteins by introducing random combinations of genes on circular DNA called plasmids. When cells are positive for a protein interaction, the researchers can use genetic sequencing to figure out what the two proteins interacting are, using new high-throughput DNA sequencing technologies similar to those used for human genome sequencing.
This way, they're no longer limited to testing one “bait” protein at a time, but could test the interactions between all the proteins in a library at once.
Ecker's group tested the new method, dubbed CrY2H-seq, on all the transcription factors–a large class of proteins–in the plant Arabidopsis.
“When you take 1,800 proteins and test the interactions among them, that's nearly 4 million combinations,” says Ecker. “We did that ten times in a matter of a month.”
They revealed more than 8,000 interactions among those proteins tested, giving them new insight into which Arabidopsis transcription factors interact with each other. The data, they say, helps answer longstanding questions about whether certain groups of transcription factors have set functions. Some of the poorly understood transcription factors, they found, interact with more well-understood factors that regulate the plant's response to auxin, a hormone involved in coordinating plant growth.
In the future, the method could be scaled up to test larger sets of proteins–human cells, for instance, contain about 20,000 different proteins. This easier and faster method to determine the entire interactome of a cell also opens up the possibility of studying how the interactome changes under different conditions–an experiment that's never been possible in the past.
###
Other researchers on the study were Renee Garza, Andrew MacWilliams, Joseph Nery, Anna Bartlett, Rosa Castanon, Adeline Goubil, Joseph Feeney, Ronan O'Malley, Shao-shan Carol Huang, Zhuzhu Zhang, and Mary Galli of the Salk Institute.
The work and the researchers involved were supported by grants from the U.S. Department of Energy, National Science Foundation Graduate Research Fellowship Program, Howard Hughes Medical Institute, and Mary K. Chapman Foundation.
About the Salk Institute for Biological Studies:
Every cure has a starting point. The Salk Institute embodies Jonas Salk's mission to dare to make dreams into reality. Its internationally renowned and award-winning scientists explore the very foundations of life, seeking new understandings in neuroscience, genetics, immunology and more. The Institute is an independent nonprofit organization and architectural landmark: small by choice, intimate by nature and fearless in the face of any challenge. Be it cancer or Alzheimer's, aging or diabetes, Salk is where cures begin. Learn more at: salk.edu.












