
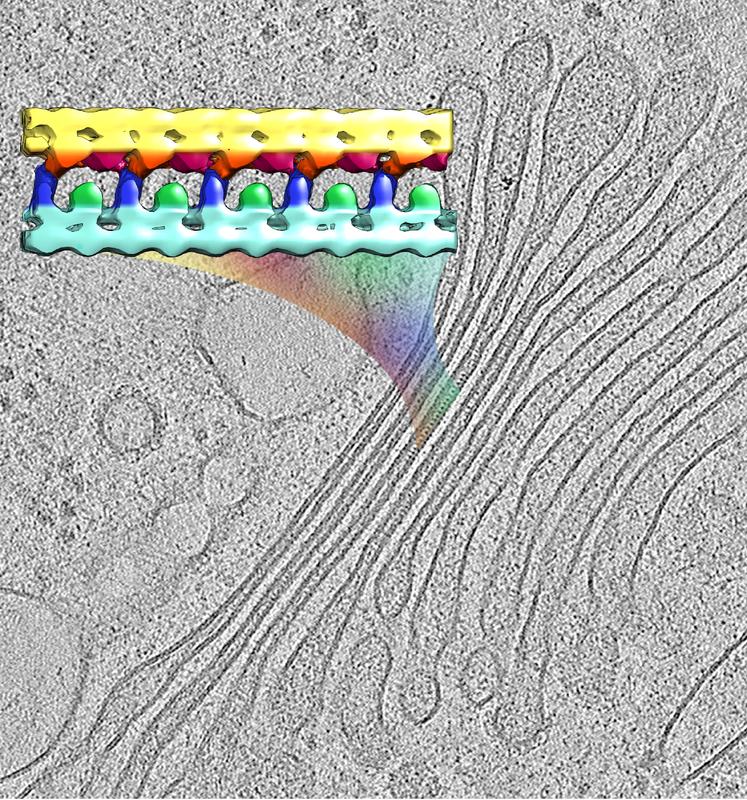
The image shows the 3D molecular structure of a membrane-linking protein array (color), extracted from a cryo-tomogram of the native Golgi (greyscale).
Sahradha Albert / Copyright: MPI of Biochemistry
It consists of flat membrane-enclosed compartments (called cisternae) that are densely packed on top of each other, like a stack of pancakes. Researchers at the Max Planck Institute of Biochemistry in Martinsried, Germany, have now identified structures within these cisternae.
“Using cryo-electron tomography, we discovered that the cisterna membranes are held together by linker proteins,” explains Benjamin Engel, first author of the study. Their results have been published in the journal PNAS.
After being produced at the endoplasmic reticulum, proteins enter the Golgi apparatus and travel through its stacks of cisterna membranes. As the proteins move through the cisternae, they receive a variety of modifications and are finally sorted into vesicles for delivery to different locations inside and outside the cell.
The elaborate membrane architecture of the Golgi is crucial for regulating the modification and sorting of cargo proteins, as different Golgi enzymes are localized to specific cisterna stacks. However, many questions remain about how the Golgi architecture is established. Researchers at the MPI of Biochemistry helped answer these questions by using in situ cryo-electron tomography to identify new molecular structures inside the Golgi of the alga Chlamydomonas.
Until recently, researchers could only use traditional electron microscopy to look closely at cellular structures. However, the sample preparation steps required for this technique can damage the specimen and thus prevent the observation of fine molecular details.
Scientists in the “Molecular Structural Biology“ Department, led by Prof. Wolfgang Baumeister, have engineered a method called in situ cryo-electron tomography. The cell is rapidly frozen to preserve its delicate structure and then thinned with a focused ion beam, revealing the cellular interior. Next, an electron microscope is used to acquire a three-dimensional view of the unperturbed molecular environment inside the cell.
Experts in the Golgi field assumed for a long time that the cisterna stacks were not held together by linker proteins. By using cryo-electron tomography to look at the Golgi apparatus, Benjamin Engel and his colleagues discovered arrays of proteins between the cisterna membranes (see figure), which had gone unseen using other techniques. “The way the protein arrays hold two Golgi membranes together is similar to how a zipper works when you put on a jacket”, explains PhD student Shoh Asano, co-author of the study.
The researchers believe that the protein arrays may have several functions to help the Golgi carry out its role as the cell’s post office. Do these structures define a sub-compartment of the Golgi that accelerates the enzyme reactions used to modify cargo proteins? Do the protein arrays physically force larger cargo proteins to the periphery of the Golgi, where they are sorted into vesicles for delivery? These are questions that Benjamin Engel and his colleagues want to solve in the future.
Original publication:
B. D. Engel, M. Schaffer, S. Albert, S. Asano, J. M. Plitzko and W. Baumeister: In situ structural analysis of Golgi intracisternal protein arrays. Proceedings of the National Academy of Sciences USA, September 8, 2015
Doi: 10.1073/pnas.1515337112
Contact
Prof. Dr. Wolfgang Baumeister
Molecular Structural Biology
Max Planck Institute of Biochemistry
Am Klopferspitz 18
82152 Martinsried
Germany
E-Mail: baumeist@biochem.mpg.de
http://www.biochem.mpg.de/baumeister
Anja Konschak
Public Relations
Max Planck Institute of Biochemistry
Am Klopferspitz 18
82152 Martinsried
Germany
Tel. +49 89 8578-2824
E-Mail: konschak@biochem.mpg.de
http://www.biochem.mpg.de
http://www.biochem.mpg.de/en/news – More press releases of the MPI of Biochemistry
http://www.biochem.mpg.de/baumeister – Website of the Research Department “Molecular Structural Biology” (Wolfgang Baumeister)












