Getting a grip on slippery cell membranes
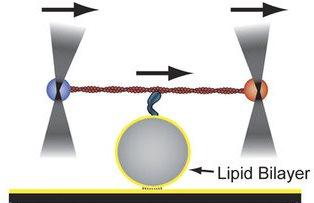
To study the forces generated by myosin-1 motors on oily membranes, researchers at Worcester Polytechnic Institute (WPI) and the University of Pennsylvania (Penn) strung a motor attached to an actin filament between two fluorescent beads, with the motor's tail resting on a sphere covered with lipids. The filament was moved side-to-side with the help of an optical trap. As the myosin-1 molecules stretched and slipped on the sphere, the researchers measured the forces applied to the sphere by the molecule. Credit: Worcester Polytechnic Institute and University of Pennsylvania
Within each of our cells is a distribution system that uses molecular motors and filaments to move proteins, organelles, and other tiny bits of cargo along its inner framework, or cytoskeleton.
To achieve this feat, the motors and filaments must tug on flexible membranes that surround the cargo packages, but these membranes, made of fatty molecules called lipids, are extremely slippery. Scientists have long wondered how the molecular transport machinery is able to maintain its grip.
The work is important because knowledge of the basic science of molecular motors and membrane mechanics can translate into a better understanding of cell and tissue development, wound healing, and the responses of the immune system–and how cancer cells can spread from a single tumor to other areas of the body.
Thanks to a collaborative research project at Worcester Polytechnic Institute (WPI) and the University of Pennsylvania (Penn), the answer is beginning to emerge. Led by Erkan Tüzel, PhD, associate professor of physics at WPI, and Michael Ostap, PhD, professor of physiology at the Pennsylvania Muscle Institute and Penn's Perelman School of Medicine, the team is using laboratory experiments and computational modeling to study the interactions between the motors (made from a protein called myosin-1), the filaments (made from the protein actin), and the membranes. Their findings are reported in the paper “Force Generation by Membrane-Associated Myosin-1” published online by Nature Scientific Reports.
“To maintain a grip, these myosin-1 molecules need to generate sufficient force against oily membranes,” Tüzel said. “How they do that has not been clear. Now we are able to say, 'yes, the numbers make sense and the physics does work.'”
In addition to transporting cargo, cells perform a number of essential functions–from secreting proteins to dividing into two daughter cells–that require the precise manipulation of cellular membranes. The work is done by myosin motors attached to actin filaments, which must grip the membranes and pull against them. This work provides novel insights into how motors keep their grip.
Getting to the bottom of this mystery required the combined skills of Tüzel, a theoretical biophysicist who develops algorithms and computational models that simulate the behavior of complex systems, including living cells, and Ostap, an experimental biophysicist who studies the molecular motors and other structures that power cells. They began collaborating after meeting in 2014 at the Muscle and Molecular Motors Gordon Research Conference.
For the current study, Serapion Pyrpassopoulos, PhD, a researcher in Ostap's lab, strung myosin-1 molecules attached to an actin filament between two fluorescent beads. The tail of the myosin was placed on a sphere covered with lipids using techniques developed by Pyrpassopoulos.
The actin filament was moved side-to-side with the help of an optical trap. As the myosin-1 molecules stretched and slipped on the sphere, the researchers measured the forces applied to the sphere by the molecule. Tüzel and WPI graduate student Göker Arpag? took the data from those single-molecule experiments and developed a computational model that could be used to determine what it would take for myosin-1 molecules to effectively manipulate a membrane.
The model showed that a single myosin-1 molecule gripping a single lipid molecule in the membrane is not able to generate the force required to successfully tug on the membrane. In fact, the model predicts, it would take between 69 and 124 myosin-1 molecules, all attached to one actin filament and all working together, to do the job. The model also predicts that myosin-1 molecules will slip on the membrane's lipid surface at different rates. When some find an area that is easier to grip, others migrate there and hold on collectively, much like a tug-of-war team that bunches up on the rope where it gains a good foothold.
“We also saw that the slower moving myosins seem to help the faster slipping ones by giving them time to move to the area where it was easier to grip,” Tüzel noted.
Tüzel and Ostap are continuing their collaboration, planning new experiments based on the predictions of the computer model. “These basic experiments and models are exciting because they provide us with the framework to start asking more physiologically relevant questions,” Ostap added, “like what happens to myosin's force-generating properties when it attaches to its cellular cargo.”
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















