
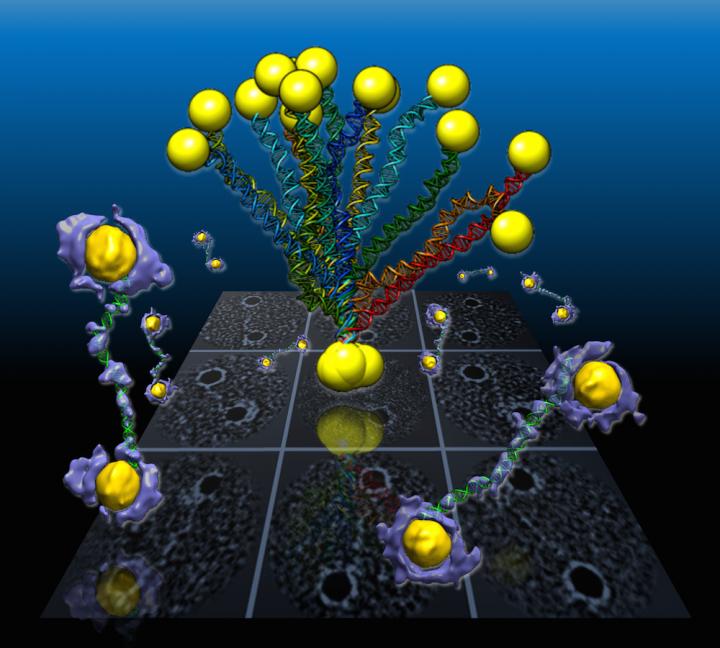
In a Berkeley Lab-led study, flexible double-helix DNA segments connected to gold nanoparticles are revealed from the 3-D density maps (purple and yellow) reconstructed from individual samples using a Berkeley Lab-developed technique called individual-particle electron tomography or IPET. Projections of the structures are shown in the background grid.
Credit: Berkeley Lab
An international team working at the Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) has captured the first high-resolution 3-D images from individual double-helix DNA segments attached at either end to gold nanoparticles. The images detail the flexible structure of the DNA segments, which appear as nanoscale jump ropes.
This unique imaging capability, pioneered by Berkeley Lab scientists, could aid in the use of DNA segments as building blocks for molecular devices that function as nanoscale drug-delivery systems, markers for biological research, and components for computer memory and electronic devices. It could also lead to images of important disease-relevant proteins that have proven elusive for other imaging techniques, and of the assembly process that forms DNA from separate, individual strands.
The shapes of the coiled DNA strands, which were sandwiched between polygon-shaped gold nanoparticles, were reconstructed in 3-D using a cutting-edge electron microscope technique coupled with a protein-staining process and sophisticated software that provided structural details to the scale of about 2 nanometers, or two billionths of a meter.
“We had no idea about what the double-strand DNA would look like between the nanogold particles,” said Gang “Gary” Ren, a Berkeley Lab scientist who led the research. “This is the first time for directly visualizing an individual double-strand DNA segment in 3-D,” he said. The results were published in the March 30 edition of Nature Communications.
The method developed by this team, called individual-particle electron tomography (IPET), had earlier captured the 3-D structure of a single protein that plays a key role in human cholesterol metabolism. By grabbing 2-D images of the same object from different angles, the technique allows researchers to assemble a 3-D image of that object. The team has also used the technique to uncover the fluctuation of another well-known flexible protein, human immunoglobulin 1, which plays a role in our immune system.
For this latest study of DNA nanostructures, Ren used an electron-beam study technique called cryo-electron microscopy (cryo-EM) to examine frozen DNA-nanogold samples, and used IPET to reconstruct 3-D images from samples stained with heavy metal salts. The team also used molecular simulation tools to test the natural shape variations, called “conformations,” in the samples, and compared these simulated shapes with observations.
Ren explained that the naturally flexible dynamics of samples, like a man waving his arms, cannot be fully detailed by any method that uses an average of many observations.
A popular way to view the nanoscale structural details of delicate biological samples is to form them into crystals and zap them with X-rays, though this does not preserve their natural shape and the DNA-nanogold samples in this study are incredibly challenging to crystallize. Other common research techniques may require a collection of thousands near-identical objects, viewed with an electron microscope, to compile a single, averaged 3-D structure. But this 3-D image may not adequately show the natural shape fluctuations of a given object.
The samples in the latest experiment were formed from individual polygon gold nanostructures, measuring about 5 nanometers across, connected to single DNA-segment strands with 84 base pairs. Base pairs are basic chemical building blocks that give DNA its structure. Each individual DNA segment and gold nanoparticle naturally zipped together with a partner to form the double-stranded DNA segment with a gold particle at either end.
The samples were flash-frozen to preserve their structure for study with cryo-EM imaging, and the distance between the two gold particles in individual samples varied from 20-30 nanometers based on different shapes observed in the DNA segments. Researchers used a cryo-electron microscope at Berkeley Lab's Molecular Foundry for this study.
They collected a series of tilted images of the stained objects, and reconstructed 14 electron-density maps that detailed the structure of individual samples using the IPET technique. They gathered a dozen conformations for the samples and found the DNA shape variations were consistent with those measured in the flash-frozen cryo-EM samples. The shapes were also consistent with samples studied using other electron-based imaging and X-ray scattering methods, and with computer simulations.
While the 3-D reconstructions show the basic nanoscale structure of the samples, Ren said that the next step will be to work to improve the resolution to the sub-nanometer scale.
“Even in this current state we begin to see 3-D structures at 1- to 2-nanometer resolution,” he said. “Through better instrumentation and improved computational algorithms, it would be promising to push the resolution to that visualizing a single DNA helix within an individual protein.”
The technique, he said, has already excited interest among some prominent pharmaceutical companies and nanotechnology researchers, and his science team already has dozens of related research projects in the pipeline.
In future studies, researchers could attempt to improve the imaging resolution for complex structures that incorporate more DNA segments as a sort of “DNA origami,” Ren said. Researchers hope to build and better characterize nanoscale molecular devices using DNA segments that can, for example, store and deliver drugs to targeted areas in the body.
“DNA is easy to program, synthesize and replicate, so it can be used as a special material to quickly self-assemble into nanostructures and to guide the operation of molecular-scale devices,” he said. “Our current study is just a proof of concept for imaging these kinds of molecular devices' structures.”
###
The Molecular Foundry is a DOE Office of Science User Facility.
In addition to Berkeley Lab scientists, other researchers contributing to this study were from UC Berkeley, the Kavli Energy NanoSciences Institute at Berkeley Lab and UC Berkeley, and Xi'an Jiaotong University in China.
This work was supported by the National Science Foundation, DOE Office of Basic Energy Sciences, National Institutes of Health, the National Natural Science Foundation of China, Xi'an Jiaotong University in China, and the Ministry of Science and Technology in China.
View more about Gary Ren's research group here.
Lawrence Berkeley National Laboratory addresses the world's most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab's scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy's Office of Science. For more, visit http://www.
DOE's Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit http://science.












