Understanding enzyme evolution paves the way for green chemistry
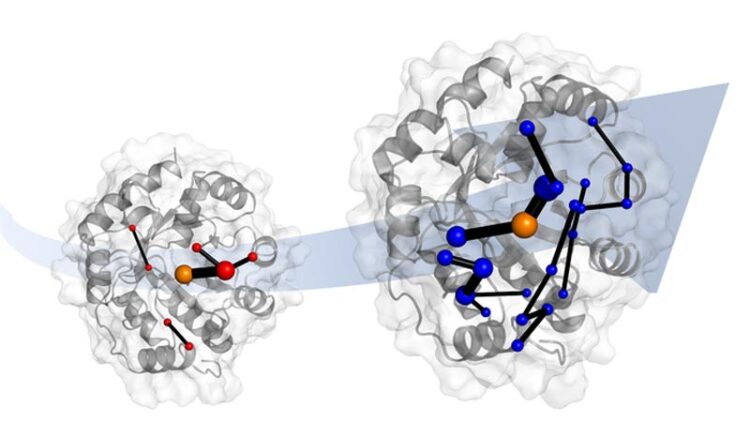
Laboratory evolution of a designer enzyme makes it into a much better catalyst . Simulations show that evolution does this by introducing networks of amino acids. These networks promise to be templates for catalyst design.
Credit: Dr. Adrian Bunzel
Researchers at the University of Bristol have shown how laboratory evolution can give rise to highly efficient enzymes for new-to-nature reactions, opening the door for novel and more environmentally friendly ways to make drugs and other chemicals.
Scientists have previously designed protein catalysts from scratch using computers, but these are much less capable than natural enzymes. To improve their performance, a technique called laboratory evolution can be used, which American chemical engineer Frances Arnold pioneered and for which she received the Nobel Prize in 2018. Directed evolution imitates natural selection, allowing scientists to use the power of biology to improve the ability of proteins to carry out tasks such as catalysing a specific chemical reaction.
But although the research team had recently used laboratory evolution to improve a designed enzyme by more than 1,000 fold, it was unknown how evolution boosts its activity. Until now.
Lead author Professor Adrian Mulholland of Bristol’s School of Chemistry said: “Evolution can make catalysts much more active. The thing is, evolution works in mysterious ways: for example, mutations that apparently improve catalysis often involve changes in amino acids far from the active site where the reaction happens.”
“We wanted to understand how evolution can transform inefficient designer biocatalysts into highly active enzymes.”, the first author of the study, Dr Adrian Bunzel, said.
To do so, the international research team from Bristol, the ETH Zurich and the University of Waikato (NZ) turned to molecular computer simulations. “These show that evolution changes the way the protein moves – its dynamics. Put simply, evolution ‘tunes’ the flexibility of the whole protein,” he added.
The team also identified the network of amino acids in the protein responsible for this ‘tuning’. These networks involve parts of the protein that are changed by evolution.
Dr Bunzel said: “After evolution, the whole protein seems to work together to accelerate the reaction. This is important because when we design enzymes, we often focus only on the active site only, and forget about the rest of the protein.”
Prof Mulholland added: “This sort of analysis could help to design more effective ‘de novo’ enzymes, for reactions that previously we could not target.”
The research, published in Nature Chemistry, reveals how evolution makes designer enzymes more powerful, paving the way to tailor-made catalysts for green chemistry.
The researchers will now use their findings to help design new protein catalysts.
Paper:
‘Evolution of dynamical networks enhances catalysis in a designer enzyme’ in Nature Chemistry by HA Bunzel, JLR Anderson, D Hilvert, VL Arcus, MW van der Kamp and AJ Mulholland
Notes to Editor
With thanks to:
EPSRC (EP/M013219/1 and EP/M022609/1)
BBSRC (BB/R016445/1)
Swiss National Science Foundation (Postdoc. Mobility fellowship)
Marsden Fund of New Zealand (16-UOW-027).
This work was conducted using the computational facilities of the Advanced Computing Research Centre, University of Bristol.
Journal: Nature Chemistry
Method of Research: Computational simulation/modeling
Subject of Research: Not applicable
Article Title: Evolution of dynamical networks enhances catalysis in a designer enzyme
Article Publication Date: 18-Aug-2021
Media Contact
Laura Thomas
lauram.thomas@bristol.ac.uk
Office: 07-977-983-814
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Bringing bio-inspired robots to life
Nebraska researcher Eric Markvicka gets NSF CAREER Award to pursue manufacture of novel materials for soft robotics and stretchable electronics. Engineers are increasingly eager to develop robots that mimic the…

Bella moths use poison to attract mates
Scientists are closer to finding out how. Pyrrolizidine alkaloids are as bitter and toxic as they are hard to pronounce. They’re produced by several different types of plants and are…

AI tool creates ‘synthetic’ images of cells
…for enhanced microscopy analysis. Observing individual cells through microscopes can reveal a range of important cell biological phenomena that frequently play a role in human diseases, but the process of…





















