New Method Refines Cell Sample Analysis
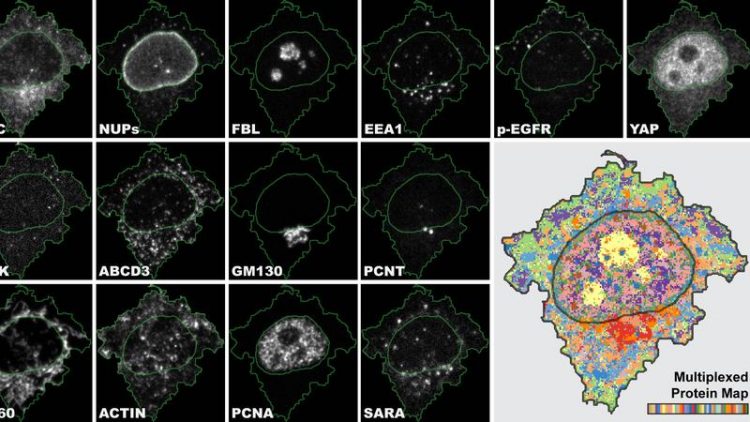
4i is the first imaging technique which gives us a multiplexed tissue-to-organelle view of biological samples and links multiple information in one and the same experiment. UZH
Researchers at the University of Zurich have developed a novel method for analyzing cells and their components called Iterative Indirect Immunofluorescence Imaging (4i). This innovation greatly refines the standard immunofluorescence imaging technique used in biomedicine and provides clinicians with an enormous amount of data from each individual sample.
4i makes it possible to observe the spatial distribution of at least 40 proteins and their modifications in the same cell for hundreds of thousands of cells simultaneously at various levels, from the tissue down to the organelle level.
Ten times more proteins visualized at the same time
“4i is the first imaging technique which gives us a multiplexed tissue-to-organelle view of biological samples. We can, for the first time, link multiplexed information derived at the tissue, cellular and subcellular level in one and the same experiment,” says Gabriele Gut, lead author of the study and postdoctoral researcher at the Institute of Molecular Life Sciences at UZH.
Immunofluorescence (IF) uses antibodies to visualize and locate proteins in biological samples. While the standard IF method usually marks three proteins, 4i uses off-the-shelf antibodies and conventional fluorescence microscopes to visualize ten times more proteins by iterative hybridization and removal of antibodies from the sample.
“Imagine cell biologists to be journalists. Every experiment is an interview with our cells. With conventional IF I can ask three questions, whereas with 4i I can have a discussion on more than 40 topics,” explains Gabriele Gut.
Map provides systematic survey of cellular landscape
Once acquired, the huge amount of data must then also be able to be analyzed – the next hurdle for the researchers. “We generated images with subcellular resolution for thousands of cells for 40 channels for more than 10 treatment conditions. The human eye and brain cannot process the biological complexity collected by 4i”.
To make full use of the 4i data, Gabriele Gut developed a new computer program for visualization and analysis called Multiplexed Protein Maps. It extracts the multiplexed fluorescence signal for millions of pixels and generates an abstract but representative map of the multiplexed protein distribution in cells.
The researchers were thus able to generate a systematic survey of the cellular landscape: They managed to visualize the spatial intracellular organization of most mammalian organelles along the cell cycle and in different microenvironments.
Advancing precision medicine
The applications for 4i and multiplexed protein maps are manifold, ranging from basic research to precision medicine. “We hope that 4i and multiplexed protein maps will help researchers to understand processes better that have been at the center of biological research for decades,” says Gut. At the same time, the researchers plan to use these technologies to advance the frontiers of precision medicine, particularly in cancer diagnosis and therapy selection.
New method already being applied in tumor therapy
The new Iterative Indirect Immunofluorescence Imaging (4i) analysis method can also be used to determine the effects of pharmacological substances on the organization and physiology of cells. It is currently being used in a translational research collaboration with clinicians and a pharmaceutical company with the aim of improving treatment outcome of cancer patients. Lucas Pelkmans, professor at the Institute of Molecular Life Sciences at UZH, and his research team aim to characterize tumor cells of patients who have been treated with different cancer medications.
The scientists hope that the lab results will provide information to support clinical decision-making for the individual treatment of patients. Moreover, the researchers plan to implement 4i and multiplexed protein maps on tissue sections of tumors to identify relevant biomarkers and thus improve diagnoses and prognoses for cancer patients.
Gabriele Gut, PhD
Institute of Molecular Life Sciences
University of Zurich
Phone: +41 44 635 31 53
E-mail: gabriele.gut@uzh.ch
Gabriele Gut, Markus D. Herrmann, Lucas Pelkmans. Multiplexed protein maps link subcellular organization to cellular states. Science, 3 August 2018. DOI: 10.1126/science.aar7042
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Combatting disruptive ‘noise’ in quantum communication
In a significant milestone for quantum communication technology, an experiment has demonstrated how networks can be leveraged to combat disruptive ‘noise’ in quantum communications. The international effort led by researchers…
Stretchable quantum dot display
Intrinsically stretchable quantum dot-based light-emitting diodes achieved record-breaking performance. A team of South Korean scientists led by Professor KIM Dae-Hyeong of the Center for Nanoparticle Research within the Institute for…

Internet can achieve quantum speed with light saved as sound
Researchers at the University of Copenhagen’s Niels Bohr Institute have developed a new way to create quantum memory: A small drum can store data sent with light in its sonic…





















